Difference between revisions of "2017 DOCK tutorial 1 with PDB 4QMZ NEW"
Stonybrook (talk | contribs) (→III. Generating Receptor Surface and Spheres) |
Stonybrook (talk | contribs) (→III. Generating Receptor Surface and Spheres) |
||
| Line 149: | Line 149: | ||
The image that appears should resemble this: | The image that appears should resemble this: | ||
| − | [[ | + | [[Image:4qmz_spheres.png|thumb|center|375px|4qmz receptor with all spheres]] |
| Line 155: | Line 155: | ||
Run | Run | ||
| − | sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 | + | sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 10.0 |
This command will select all of the spheres within 8.0 angstroms of the ligand and output them to selected_spheres.sph | This command will select all of the spheres within 8.0 angstroms of the ligand and output them to selected_spheres.sph | ||
| Line 165: | Line 165: | ||
Actions -> Atoms/Bonds -> sphere | Actions -> Atoms/Bonds -> sphere | ||
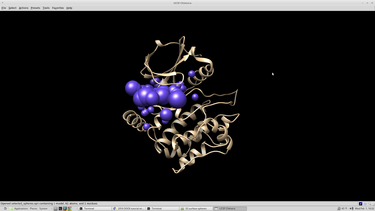
The selected spheres with the receptor surface should look similar to that as seen below: | The selected spheres with the receptor surface should look similar to that as seen below: | ||
| + | |||
| + | [[Image:Selectedspheres.png|thumb|center|375px|4qmz receptor surface with the selected spheres within 10.0 Angstroms]] | ||
==IV. Generating Box and Grid== | ==IV. Generating Box and Grid== | ||
Revision as of 18:07, 6 February 2017
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by a subsection of the AMS 536 class of 2017, using DOCK v6.8.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
4QMZ
In this tutorial we will use PDB code 4QMZ, the deposited crystal structure of MST3 in complex with SUNITINIB.
Organizing Directories
While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify.For this tutorial, we will use something similar to the following:
~username/AMS536-Spring2016/dock-tutorial/00.files/
/01.dockprep/
/02.surface-spheres/
/03.box-grid/
/04.dock/
/05.large-virtual-screen/
/06.virtual-screen/
/07.footprint/
/08.print_fps
|
In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '4QMZ'. The following sections in this tutorial will adhere to this directory structure/naming scheme.
II. Preparing the Receptor and Ligand
Download the PDB file (4QMZ)
4QMZ was moved into 00.files
4qmz.pdb was copied to raw_4qmz.pdb
raw_4qmz.pdb was opened with VI terminal editor
The header information, connect records, ions (atoms 2333 and 2334) and waters were deleted
Res 178 = TPO, or phosphonothreonine
Res 178 (TPO) was renamed to THR (Threonine) and HETATM renamed to ATOM, in addition the acanonical atoms were removed from the pdb leaving a deprotonated threonine (Atoms 1311-1314 in 4qmz.pdb)
Res B49 was renamed to LIG and made Chain B
raw_4qmz.pdb was copied twice to 4qmz_rec.pdb and 4qmz_lig.pdb
4qmz_rec.pdb was opened with VI terminal editor
LIG atoms, or chain B, was deleted and the file saved
4qmz_lig.pdb was opened with VI terminal editor
Protein atoms, or chain A, was deleted and the file saved
4qmz_rec.pdb was loaded into tleap as a quality control measure
tleap
source leaprc.protein.ff14SB
lin = loadpdb /path/to/4qmz_rec.pdb
2340 Hydrogens added, 1 heavy atom added (CSER RES 299, Chain A, OXT 12)
check lin
saveamberparm lin /path/to/4qmz_rec_leap.parm7 /path/to/4qmz_rec_leap.crd
Running the receptor through leap ensures a reasonable starting structure and can help identify obvious issues sooner rather than later.
At this point the .parm7 and .crd have been created via tleap ambpdb can be used to obtain the clean pdb 4qmz_rec_leap.pdb
ambpdb -p 4qmz_rec_leap.parm7 -c 4qmz_rec_leap.crd > 4qmz_rec_leap.pdb
Now add partial charges to the receptor and save file in .mol2 format:
open chimera
load 4qmz_rec_leap.pdb
Tools --> Structure editing --> Add charge --> AMBER ff99SB with AM1-BCC charges
File --> Save Mol2... --> 4qmz.rec.mol2
A no-hydrogen receptor pdb file will now be created:
Chimera, load 4qmz.rec.mol2
Select --> Chemistry --> element --> H
Actions --> Atoms --> Delete
File --> Save PDB... --> 4qmz.rec.noH.pdb
Ligand (4qmz_lig.pdb) will now be charged and saved in mol2 format
open chimera
load 4qmz_lig.pdb
Tools --> structure editing --> AddH
Tools --> Structure editing --> Add charge --> AMBER ff99SB with AM1-BCC charges
File --> Save mol2 --> 4qmz.lig.mol2
Placement of partial charges can be verified by examining the saved files 4qmz.lig.mol2 and 4qmz.rec.mol2.
III. Generating Receptor Surface and Spheres
Open 4qmz.rec.noH.pdb in Chimera
To generate the molecular surface:
Action --> Surface --> Show
Save the .dms file
Tools --> Structure editing --> write DMS
.dms save to 4qmz.rec.noH.dms
Create surface spheres
create input file INSPH
4qmz.rec.noH.dms
R
X
0.0
4.0
1.4
4qmz.rec.sph
line 1 designates input file line 2 designates the generated spheres will be outside the receptor surface line 3 designates that all points on the receptor will be used line 4 designates the maximum surface radius of the spheres line 5 designates the minimum surface radius of the spheres line 6 designates the output file name
run sphgen
sphgen -i INSPH -o OUTSPH
sphgen is the sphere generation program from dock -i desginates the input file: INSPH -o designates the output file
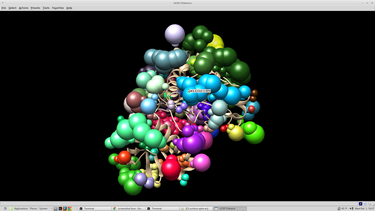
At this point it is beneficial to visualize the spheres that were created. This can be done with chimera: Open chimera from the terminal choose file --> open 4qmz.rec.mol2 choose file --> open 4qmz.rec.sph
The image that appears should resemble this:
Then we need to select the spheres pertinent to our docking experiment. Usually these speheres will be the closest N spheres to the native ligand molecule.
Run
sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 10.0
This command will select all of the spheres within 8.0 angstroms of the ligand and output them to selected_spheres.sph
To visualize the spheres using Chimera as previously done: Launch Chimera, choose File -> Open, choose 4qmz.rec.noH.pdb File -> Open, choose output_spheres_selected.pdb Select -> Residue -> SPH Actions -> Atoms/Bonds -> sphere The selected spheres with the receptor surface should look similar to that as seen below:
IV. Generating Box and Grid
enter directory 03.box-grid
create input showbox.in
Y
8.0
../02.surface-spheres/selected_spheres.sph
1
4qmz.box.pdb
This input designates to dock that: we want to create a box the box length should be 8.0 Angstroms use the selected spheres in the file desginated output the box to the file specified
to use this input type:
showbox < showbox.in
This box can be visualized in chimera using a similar approach as visualizing the spheres
Compute the energy grid
create grid.in file
vi grid.in
grid.in needs to contain:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 12 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../01.dockprep/4qmz.rec.mol2 box_file ../03.box-grid/4qmz.box.pdb vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
this script should output verbosely to the terminal and produce 2 output files, grid.bmp and grid.nrg, both binary files.
V. Docking a Single Molecule for Pose Reproduction
Create or enter directory 4, 04.dock
Minimization
create min.in
vi min.in
or
touch min.in
min.in should contain:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01.dockprep/4qmz.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
run:
dock6 -i min.in
this command will output 4qmz.lig.min_scored.mol2
This structure can be visualized using chimera and loading the surface into chimera along with the minimized and unminimized ligand structures:
At this point we are going to calculate the van der wals and electrostatic footprints for the unminimized and minimized ligand structures in relation to the receptor active site.
create the input file: footprint.in
conformer_search_type rigid use_internal_energy no ligand_atom_file ./4qmz.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_use_footprint_reference_mol2 yes fps_footprint_reference_mol2_filename ../01.dockprep/4qmz.lig.mol2 fps_foot_compare_type Euclidean fps_normalize_foot no fps_foot_comp_all_residue yes fps_receptor_filename ../01.dockprep/4qmz.rec.mol2 fps_vdw_att_exp 6 fps_vdw_rep_exp 12 fps_vdw_rep_rad_scale 1 fps_use_distance_dependent_dielectric yes fps_dielectric 4.0 fps_vdw_fp_scale 1 fps_es_fp_scale 1 fps_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix fps.min.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
this footprint calculation can be run in the same way as the minimization:
dock6 -i footprint.in
This will output three files: fps.min.output_scored.mol2
fps.min.output_scored_footprint_scored.txt
fps.min.output_hbond_scored.txt
With an in house script, plot_footprint_single_magnitude.py the two sets of footprints will be plotted for comparison
this script will be run using:
python plot_footprint_single_magnitude.py fps.min.output_scored_footprint_scored.txt 50
The output will look something like this: