Difference between revisions of "2018 Denovo design tutorial 2 with PDB 1C87"
| Line 140: | Line 140: | ||
| − | ==Denovo Rescore == | + | ===Denovo Rescore=== |
Create a directory for denovo rescore. | Create a directory for denovo rescore. | ||
mkdir denovo_rescore | mkdir denovo_rescore | ||
Revision as of 16:55, 23 March 2018
Contents
2018 Denovo design tutorial 1 with PDB 1C87 ( Focused )
Preparing The Files
First of all, we need to create a directory for the fragment library.
mkdir fraglib
Move to fraglib directory and create an input file, fraglib.in
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy no ligand_atom_file /gpfs/projects/AMS536/2018/seung/001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix fraglib write_orientations no num_scored_conformers 1 rank_ligands no
fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_scored.mol2, fraglib_sidechain.mol2 and fraglib_torenv.dat will be created. You can check how many linkers and side chains are in your system.
[seshin@login fraglib]$ grep -wc MOLECULE fraglib_linker.mol2 3 [seshin@login fraglib]$ grep -wc MOLECULE fraglib_sidechain.mol2 1
There are 3 linkers and 1 side chain. Once fraglib output files are created, makedenovo.in input file and run it interactively.
conformer_search_type nova dn_fraglib_scaffold_file fraglib_scaffold.mol2 dn_fraglib_linker_file fraglib_linker.mol2 dn_fraglib_sidechain_file fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file anchor1.mol2 dn_use_torenv_table yes dn_torenv_table fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 4 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_output_prefix 1c87.final use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Create vi submit.sh in order to submit a job to create the output file.
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=24 #PBS -N 1c87_denovo #PBS -V #PBS -q long echo "Job started on" date cd $PBS_O_WORKDIR ../../../../zzz.programs/dock6_new/bin/dock6 -i denovo.in -o denovo.out echo "Job finished on" date
Denovo Rescore
Create a directory for denovo rescore.
mkdir denovo_rescore
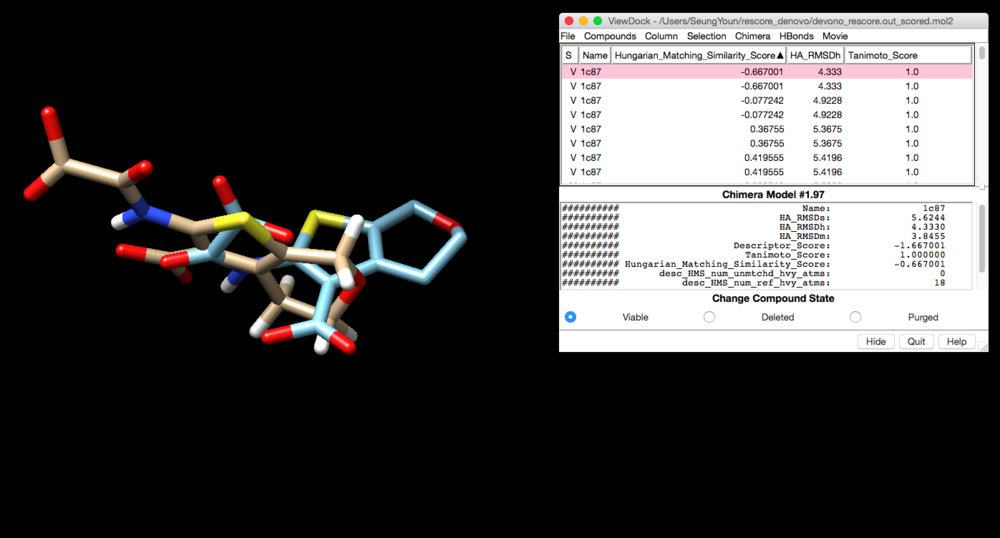
Make sure you have 1c87.final.denovo_build.mol2 file in your directory and lig_withH_charge_1c87.mol2 file will be your reference. (NOTE: Tantamoto is a chemical similarity. 0 means identical to the reference. )
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 1c87.final.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity no descriptor_use_pharmacophore_score no descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap no descriptor_fingerprint_ref_filename ../../001.files/lig_withH_charge_1c87.mol2 descriptor_hms_score_ref_filename ../../001.files/lig_withH_charge_1c87.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix devono_rescore.out write_orientations no num_scored_conformers 1 rank_ligands no
devono_rescore.out_scored.mol2 will be created. Open Chimera and load lig_withH_charge_1c87.mol2 file and viewdock to open devono_rescore.out_scored.mol2