Difference between revisions of "2018 DOCK tutorial 1 with PDBID 2NNQ"
(→Flexible Docking) |
(→VI. Virtual Screen) |
||
| (19 intermediate revisions by 4 users not shown) | |||
| Line 24: | Line 24: | ||
2.surface_spheres | 2.surface_spheres | ||
3.gridbox | 3.gridbox | ||
| − | 4. | + | 4.dock |
5. | 5. | ||
| − | 6. | + | 6.footprint |
| − | 7. | + | 7.virtual_screen |
| + | 8.virtual_screen_mpi | ||
| + | 9.cartesianmin | ||
| + | 10.rescore | ||
= II. Preparation of the ligand and receptor = | = II. Preparation of the ligand and receptor = | ||
| Line 46: | Line 49: | ||
Tools -> Structure Editing -> Add H (To add Hydrogen atoms) | Tools -> Structure Editing -> Add H (To add Hydrogen atoms) | ||
Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force filed available for standard residues. Here we used AMBER ff14SB) | Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force filed available for standard residues. Here we used AMBER ff14SB) | ||
| − | Save as a mol2 file. ( | + | Save as a mol2 file. (2nnq_rec_withH.mol2) |
- If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor. | - If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor. | ||
| Line 62: | Line 65: | ||
- Action -> Surface -> Show | - Action -> Surface -> Show | ||
- Tools -> Structure Editing -> Write DMS | - Tools -> Structure Editing -> Write DMS | ||
| − | - Save the | + | - Save the 2nnq_rec_noH.dms into 3.surface_spheres folder |
| + | |||
| + | Reopen the file and make sure the surface was generated. | ||
Transfer all the folders created so far to seawulf cluster to be used in DOCK. | Transfer all the folders created so far to seawulf cluster to be used in DOCK. | ||
===Generating spheres=== | ===Generating spheres=== | ||
| − | - Go to | + | - Go to 3.surface_spheres folder |
- Create a new input file to create spheres by typing vim INSPH and type the following lines inside the file. | - Create a new input file to create spheres by typing vim INSPH and type the following lines inside the file. | ||
| Line 114: | Line 119: | ||
The box length should be 8 Angstroms | The box length should be 8 Angstroms | ||
Use the selected_spheres file in the designated location | Use the selected_spheres file in the designated location | ||
| − | The name of the file that contains generated box. | + | The name of the pdb output file that contains generated box. |
Use the following command to generate the box. | Use the following command to generate the box. | ||
| Line 176: | Line 181: | ||
read_mol_solvation no | read_mol_solvation no | ||
calculate_rmsd yes | calculate_rmsd yes | ||
| − | use_rmsd_reference_mol | + | use_rmsd_reference_mol yes |
| + | rmsd_reference_filename ../1.dockprep/2nnq_lig_withH.mol2 | ||
use_database_filter no | use_database_filter no | ||
orient_ligand no | orient_ligand no | ||
| Line 240: | Line 246: | ||
read_mol_solvation no | read_mol_solvation no | ||
calculate_rmsd yes | calculate_rmsd yes | ||
| − | use_rmsd_reference_mol | + | use_rmsd_reference_mol yes |
| + | rmsd_reference_filename 2nnq.lig.min_scored.mol2 | ||
use_database_filter no | use_database_filter no | ||
orient_ligand yes | orient_ligand yes | ||
| Line 418: | Line 425: | ||
read_mol_solvation no | read_mol_solvation no | ||
calculate_rmsd yes | calculate_rmsd yes | ||
| − | use_rmsd_reference_mol 2nnq.lig.min_scored.mol2 | + | use_rmsd_reference_mol yes |
| + | rmsd_reference_filename 2nnq.lig.min_scored.mol2 | ||
use_database_filter no | use_database_filter no | ||
orient_ligand yes | orient_ligand yes | ||
| Line 544: | Line 552: | ||
Once everything is successful these output files should be generated. (footprint.out_footprint_scored.txt, footprint.out_hbond_scored.txt, footprint.out_scored.mol2) | Once everything is successful these output files should be generated. (footprint.out_footprint_scored.txt, footprint.out_hbond_scored.txt, footprint.out_scored.mol2) | ||
| − | Use a python script to visualize the molecular footprint. The script can be accessed in the previous DOCK tutorials. Once the script is used, the molecular footprint should be similar to the image below. Notice the large deviations in energy at different amino acid residues. Those residues contribute more towards, the interaction between the ligand and therefore can be identified as important towards the binding of new ligands which can replace the original ligand in the | + | Use a python script to visualize the molecular footprint. The script can be accessed in the previous DOCK tutorials. Once the script is used, the molecular footprint should be similar to the image below. Notice the large deviations in energy at different amino acid residues. Those residues contribute more towards, the interaction between the ligand and therefore can be identified as important towards the binding of new ligands which can replace the original ligand in the PDB file. |
| + | |||
| + | [[File:2nnqfootprint.png|thumb|center|1000px| Flexible docking results for 2nnq]] | ||
| + | |||
| + | = VI. Virtual Screen = | ||
| + | Virtual screening is the method of screening a ligand library (drug-like molecules) to filter the best ligands which can bind to the binding site of a specific receptor. Here we will be using a ligand library which contains 25000 molecules to select the best ligands which can be used to replace the original ligand that the PDB file contained. | ||
| + | |||
| + | Move to the directory 7.virtual_screen. Copy the ligand library to the same directory. | ||
| + | |||
| + | Create a new input file for virtual screen. | ||
| + | touch virtual.in | ||
| + | Use the input file to perform virtual screen using DOCK6. | ||
| + | dock6 -i virtual.in | ||
| + | |||
| + | Use the following lines to answer the prompted questions. | ||
| + | conformer_search_type flex | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | write_fragment_libraries no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file small_ligand_library.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../2.surface_spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../3.boxgrid/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix virtual.out | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Since the ligand library contains 25000 molecules, it is going to take a long time to complete the virtual screen. Therefore we are going to use the mpi version of DOCK to complete the virtual screen. Therefore you can either terminate the virtual screen which is already running by pressing ctrl+c or let it run. | ||
| + | |||
| + | =VII.Virtual Screen (MPI)= | ||
| + | Until now we used the head node of the seawulf cluster. By using the mpi version of DOCK we will be using 4 processors that contain 28 nodes in each and it will complete the virtual screen quicker. | ||
| + | Move to a new directory. (8.virtual_screen_mpi) | ||
| + | Copy the input file and the ligand database file from 7.virtual_screen directory. | ||
| + | |||
| + | To submit the job to the seawulf cluster we are using a new file. (virtual.sh) | ||
| + | vim virtual.sh | ||
| + | Type the following lines in the new file. | ||
| + | #!/bin/bash | ||
| + | #PBS -l walltime=48:00:00 | ||
| + | #PBS -l nodes=4:ppn=28 | ||
| + | #PBS -q long | ||
| + | #PBS -N 2nnq.virtual | ||
| + | #PBS -V | ||
| + | cd $PBS_O_WORKDIR | ||
| + | mpirun -np 112 dock6.mpi -i virtual.in -o 2nnq.virtual.mpi.out | ||
| + | |||
| + | The virtual screen job can be submitted to the cluster using the following command. | ||
| + | qsub virtual.sh | ||
| + | |||
| + | Type the following command to check the status of the job and other jobs that you submitted via your user login. | ||
| + | qstat -u username | ||
| + | |||
| + | =VIII.Cartesian Minimization= | ||
| + | |||
| + | Here we will use the docked molecules and perform a cartesian minimization of them. | ||
| + | |||
| + | Move to the directory 8.cartesianmin. | ||
| + | |||
| + | Create a new input file for the minimization. | ||
| + | touch min.in | ||
| + | Use the input file to perform the cartesian minimization using DOCK6. | ||
| + | dock6 -i min.in | ||
| + | |||
| + | Use the following lines to answer the prompted questions. | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file 2nnq.virtualscreen_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary yes | ||
| + | continuous_score_secondary no | ||
| + | cont_score_rec_filename ../1.dockprep/2nnq.rec.charged.mol2 | ||
| + | cont_score_att_exp 6 | ||
| + | cont_score_rep_exp 12 | ||
| + | cont_score_rep_rad_scale 1 | ||
| + | cont_score_use_dist_dep_dielectric yes | ||
| + | cont_score_dielectric 4.0 | ||
| + | cont_score_vdw_scale 1 | ||
| + | cont_score_es_scale 1 | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 2nnq.virtualscreen.minimized | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Create a new submission script for the minimization. | ||
| + | touch min.sh | ||
| + | |||
| + | #!/bin/bash | ||
| + | #PBS -l walltime=48:00:00 | ||
| + | #PBS -l nodes=1:ppn=28 | ||
| + | #PBS -q long | ||
| + | #PBS -N cartesian min | ||
| + | #PBS -V | ||
| + | cd $PBS_O_WORKDIR | ||
| + | dock6 -i min.in -o min.out | ||
| + | |||
| + | =IX.Rescoring Docked Molecules= | ||
| + | We would also like to rank our docked ligands and extract the 100 best ligands (which have the most negative, lowest scores). Footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score will all be used by Dock to rescore the virtual screen. | ||
| + | |||
| + | Move to the directory 9.rescore. | ||
| + | |||
| + | Create a new input file for the rescoring. | ||
| + | touch rescore.in | ||
| + | Use the input file to perform the cartesian minimization using DOCK6. | ||
| + | dock6 -i rescore.in | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../8.cartesianmin/2nnq.virtualscreen.minimized_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score no | ||
| + | descriptor_use_multigrid_score no | ||
| + | descriptor_use_continuous_energy no | ||
| + | descriptor_use_footprint_similarity yes | ||
| + | descriptor_use_pharmacophore_score yes | ||
| + | descriptor_use_tanimoto yes | ||
| + | descriptor_use_hungarian yes | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_fps_use_footprint_reference_mol2 yes | ||
| + | descriptor_fps_footprint_reference_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 | ||
| + | descriptor_fps_foot_compare_type Euclidean | ||
| + | descriptor_fps_normalize_foot no | ||
| + | descriptor_fps_foot_comp_all_residue yes | ||
| + | descriptor_fps_receptor_filename ../1.dockprep/2nnq.rec.withH.charged.mol2 | ||
| + | descriptor_fps_vdw_att_exp 6 | ||
| + | descriptor_fps_vdw_rep_exp 12 | ||
| + | descriptor_fps_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_dielectric 4.0 | ||
| + | descriptor_fps_vdw_fp_scale 1 | ||
| + | descriptor_fps_es_fp_scale 1 | ||
| + | descriptor_fps_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename ../4.dock/2nnq.lig.min_scored.mol2 | ||
| + | descriptor_hungarian_ref_filename ../4.dock/2nnq.lig.min_scored.mol2 | ||
| + | descriptor_hungarian_matching_coeff -5 | ||
| + | descriptor_hungarian_rmsd_coeff 1 | ||
| + | descriptor_volume_reference_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 | ||
| + | descriptor_volume_overlap_compute_method analytical | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
| + | descriptor_weight_fingerprint_tanimoto -1 | ||
| + | descriptor_weight_hungarian 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn | ||
| + | pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
Latest revision as of 11:46, 12 December 2019
This tutorial contains, a step by step by approach to dock a known ligand to a known receptor.
Contents
- 1 I. Introduction
- 2 II. Preparation of the ligand and receptor
- 3 III. Generating receptor surface and spheres
- 4 IV. Generating box and grid
- 5 V. Docking a single molecule for pose reproduction
- 6 VI. Virtual Screen
- 7 VII.Virtual Screen (MPI)
- 8 VIII.Cartesian Minimization
- 9 IX.Rescoring Docked Molecules
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
2NNQ
The tutorial will be based on the PDB file 2NNQ downloaded from the PDB Database. 2NNQ is the crystal structure for a human adipocyte fatty acid binding protein in complex with ((2'-(5-ethyl-3,4-diphenyl-1H-pyrazol-1-yl)-3-biphenylyl)oxy)acetic acid.
Organization of Directories
Maintaining a clearly organized set of folders will be helpful in finding specific files, calling different files in input files and most importantly keeping track of everything you do. We would like to recommend to maintain the following set of files throughout the tutorial.
0.files
1.dockprep
2.surface_spheres
3.gridbox
4.dock
5.
6.footprint
7.virtual_screen
8.virtual_screen_mpi
9.cartesianmin
10.rescore
II. Preparation of the ligand and receptor
Download the pdb file 2NNQ from PDB database save it in 0.files folder.
Checking the structure
- Read the article related to the PDB file to understand protonation states, charges, environmental conditions and other important information regarding the receptor and the ligand. - Open the pdb file through chimera and look at the structure. Identify the main components of the model (receptor, ligand, solvent, surfactants, metal ions) - Carefully look to identify if there are any missing residues or missing loops. (This particular PDB file didn't contain any missing loops or missing residues)
Preparation of receptor
- Open the PDB file (2NNQ.pdb) via Chimera - Isolate the receptor using select tool and delete tool in Chimera. - Save the isolated receptor as a mol2 file. (2nnq_rec_noH.mol2)
- Open 2nnq_rec_noH.mol2 file again using Chimera and use the following instructions to prepare the receptor file to be used in DOCK.
Tools -> Structure Editing -> Add H (To add Hydrogen atoms)
Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force filed available for standard residues. Here we used AMBER ff14SB)
Save as a mol2 file. (2nnq_rec_withH.mol2)
- If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor.
Tools -> Structure/Binding Analysis -> DockPrep
Preparation of ligand
- Open the PDB file via Chimera. - Using Chimera, isolate the ligand, add H atoms, add charge and save it as a mol2 file by following the same steps followed for the receptor.
Once all the files are prepared make sure to save the files in 1.dockprep folder.
III. Generating receptor surface and spheres
Preparation of DMS file
- Open 2nnq_rec_noH.mol2 using chimera. - Action -> Surface -> Show - Tools -> Structure Editing -> Write DMS - Save the 2nnq_rec_noH.dms into 3.surface_spheres folder
Reopen the file and make sure the surface was generated.
Transfer all the folders created so far to seawulf cluster to be used in DOCK.
Generating spheres
- Go to 3.surface_spheres folder - Create a new input file to create spheres by typing vim INSPH and type the following lines inside the file.
2nnq_rec_noH.dms R X 0.0 4.0 1.4 2nnq_rec.sph
The first line 2nnq_rec_noH.dms specifies the input file. R indicates that spheres generated will be outside of the receptor surface. X specifies all the points will be used. 0.0 is the distance in angstroms and it will avoid steric clashes. 4.0 is the maximum surface radius of the spheres and 1.4 is the minimum radius in angstroms.The last line 2nnq_spheres.sph creates the sph file that contains clustered spheres.
Once the INSPH file is ready, type the following command to generate the spheres.
sphgen -i INSPH -o OUTSPH
Once sphgen command is successful, 2nnq_spheres.sph file will be created. Open it up using Chimera along with 2nnq_rec_noH.mol2 file. You should get a similar output like the image below.
Selecting Spheres
Here we will be selecting the spheres which defines the binding pocket of the ligand because we are trying to direct the ligand towards that binding site rather than all over the receptor. To select the spheres type the following command.
sphere_selector 2nnq_rec.sph ../1.dockprep/2nnq_lig_withH.mol2 10.0
This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph. Visualize the selected spheres using Chimera to make sure the correct spheres are selected. Notice that, spheres around the ligand binding site are kept and all the other spheres are deleted in the image below.
IV. Generating box and grid
Generating box
Move to 3.boxgrid directory Create a new file showbox.in and write the following lines in the file.
Y 8.0 ../2.surface_spheres/selected_spheres.sph 1 2nnq.box.pdb
Each of the above lines indicate that;
We intend to generate a box The box length should be 8 Angstroms Use the selected_spheres file in the designated location The name of the pdb output file that contains generated box.
Use the following command to generate the box.
showbox < showbox.in
If this step is successful, you should see a new file (2nnq.box.pdb) in 3.boxgrid folder.
Generating grid
Create a new file (grid.in)
Use the following command to generate the grid.
grid -i grid.in -o gridinfo.out
Answer the prompted questions with the answers given below. (or you can use the following lines and include them in the grid.in file before entering the above command. If you do that these questions won't be prompted again. They will be automatically answered by grid.in file created)
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 12 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../1.dockprep/2nnq_rec_withH.mol2 box_file 2nnq.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
If the command is successful, three new files will be generated. (gridinfo.out, grid.nrg, grid.bmp). Go through gridinfo.out file to make sure all the information about the receptor in the file matches with the original information of the receptor. (Eg:- Total charge, residues and their charges) If the information doesn't match, that means you have made an error in one of the steps that you followed so far.
V. Docking a single molecule for pose reproduction
Under this section, the ligand for 2nnq.pdb will be re-docked into the receptor. 3 Methods will be used to achieve this.
1. rigid docking
2. fixed anchor docking
3. flexible docking
Energy minimization
Before performing docking, here the ligand will be subjected to energy minimization in order to remove unfavorable clashes. These clashes will affect rigid docking because in rigid docking the ligand will be docked as the complete ligand, whereas in other docking methods the ligand will be broken into fragments and the ligand will be built step by step considering favorable orientations and torsion angles after each fragment addition.
Go to the directory 4.dock and a create a new file (min.in) and enter the command below.
dock6 -i min.in
Answer the prompted questions using the answers given below or include the following lines in the min.in file at before entering the above command to avoid answering the questions manually.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../1.dockprep/2nnq_lig_withH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../1.dockprep/2nnq_lig_withH.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../3.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 2nnq.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
If the process is successful a new file (2nnq.lig.min_scored.mol2) will be generated. You can compare how is it changed from the initial structure by analyzing the RMSD value generated in the file. Visualize the new mol2 file along with receptor and the initial ligand mol2 files using Chimera to see the differences.
Rigid Docking
Create an input file for rigid docking
touch rigid.in
Run dock using the created input file.
dock6 -i rigid.in
Follow a similar approach as we did for minimization to answer the prompted questions by either answering them manually using the answers in the lines below or by including the following lines in the input file before running dock.
conformer_search_type rigid use_internal_energy yes ligand_atom_file 2nnq.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 2nnq.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../3.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 1 rank_ligands no
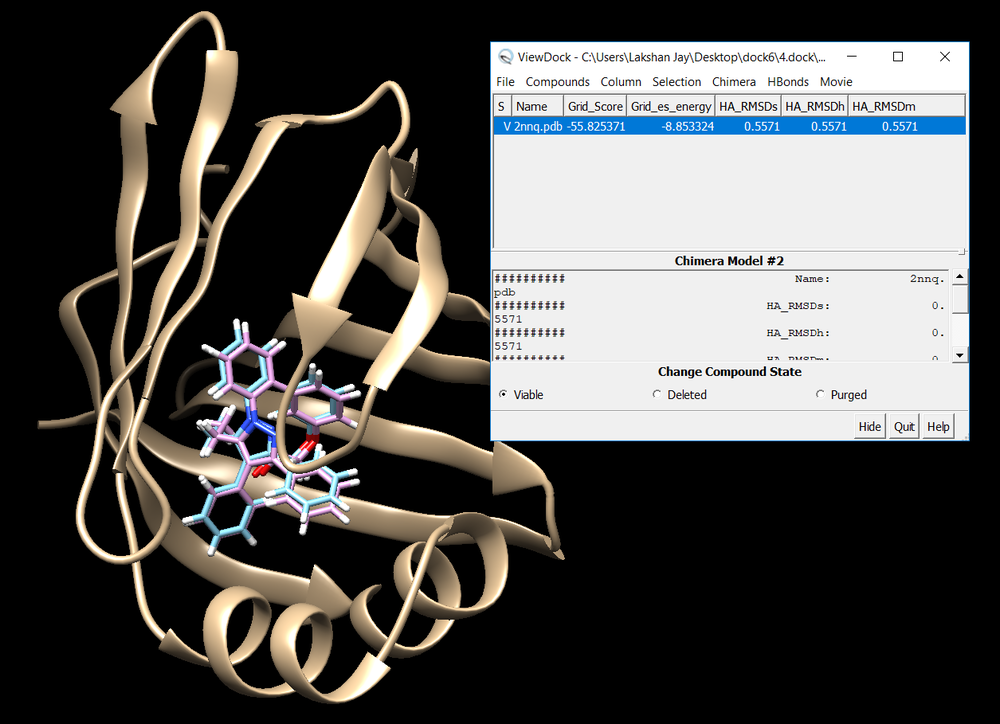
Once rigid docking is successful, you will get an output file. (rigid.out_scored.mol2) Visualize the output file using Chimera by following steps to check the rigid docking success.
Open Chimera File -> Open -> 2nnq_rec_withH.mol2 File -> Open -> 2nnq_lig_withH.mol2 Tools -> Surface/binding Analysis -> ViewDock -> Select the Rigid Dock output file. (rigid.out_scored.mol2) In the loaded dialog box select Dock4,5 or 6
Once everything is loaded go to the ViewDock window and use it's menu to view all the calculated properties regarding the rigid docked ligand by following the steps below.
Column -> Show -> gridscore Column -> Show -> HA_RMSDs Follow the same steps to get all the properties
Your visualized structure should be similar to the image below.
Fixed Anchor Docking
Create an input file for fixed anchor docking.
touch fixed.in
Use the input file to perform fixed anchor docking
dock6 -i fixed.in
Use the following lines to answer the prompted questions as we did in rigid docking.
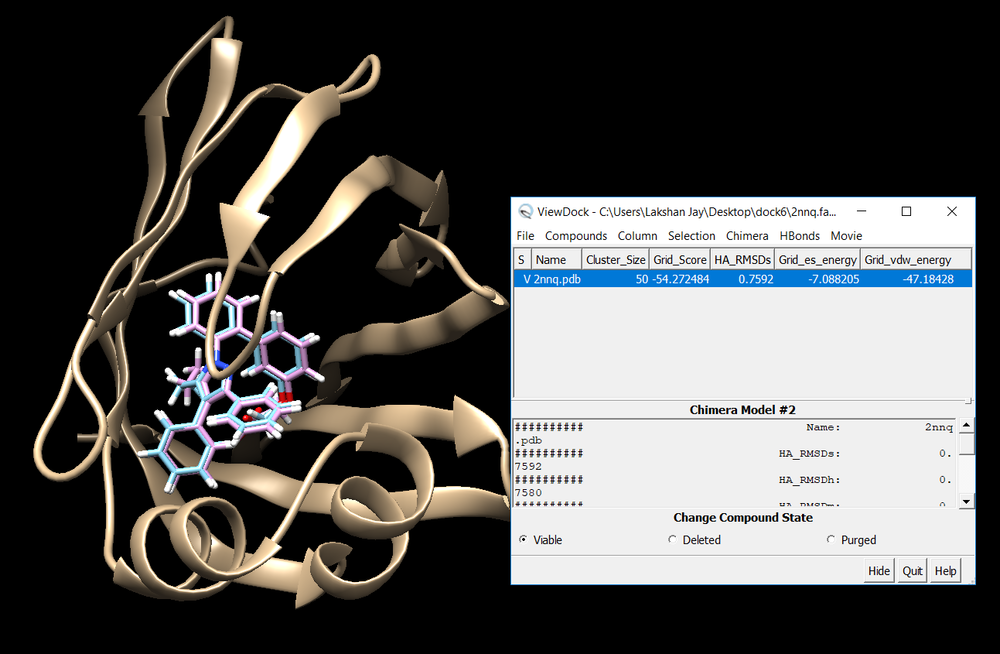
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../1.dockprep/2nnq_lig_withH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../1.dockprep/2nnq_lig_withH.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../2.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 2nnq_fad write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Once docking is completed an output file will be generated. (2nnq_fad_scored.mol2) Follow the same method used in rigid docking to visualize the docked poses using Chimera. Once it is visualized, it should like the image below. Notice all the poses 50 generated are in the same cluster and standard RMSD is 0.75. These indicate that docking is very successful.
Flexible Docking
Create a new input file for flexible docking. (flex.in)
touch flex.in
Use the created input file to perform flexible docking using DOCK6.
dock6 -i flex.in
Answer the prompted questions using the following lines as we did in rigid and fixed anchor docking.
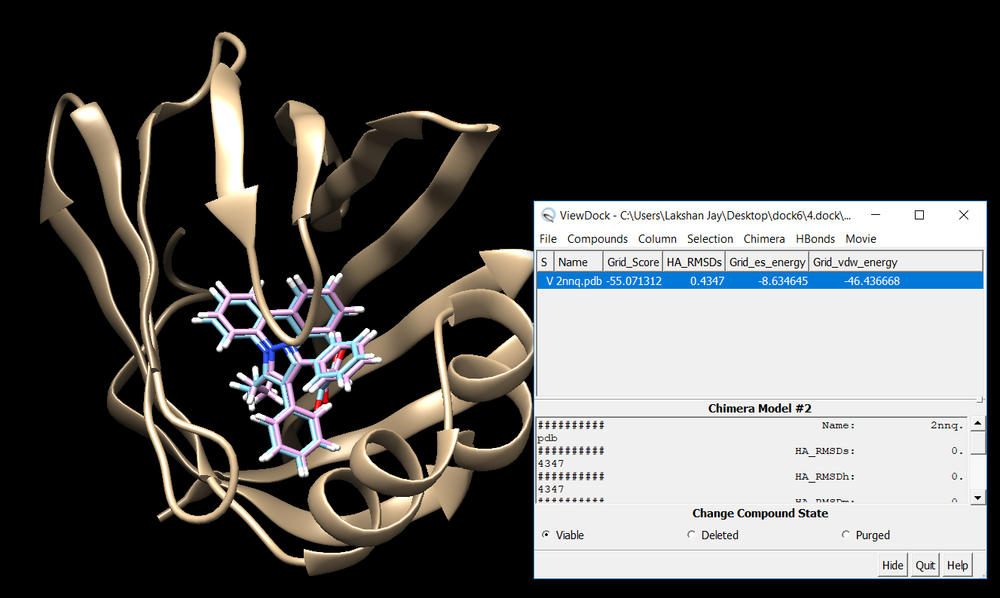
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 2nnq.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 2nnq.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../3.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 1 rank_ligands no
Once flexible docking is completed an output mol2 file will be generated. (flex.out_scored.mol2). Use the visualization steps used in rigid and fixed anchor docking and study the properties of the docking results.
Molecular Footprint
Molecular footprints can be used to determine how a ligand interacts with the receptor. Usually, the molecular footprint shows electrostatic interactions and Van der Waals interactions. Here, the molecular footprint will be used to determine how the ligand interacts with the receptor before and after minimization. To generate molecular footprints use following steps.
Go to directory 6.footprint
Generate an input file by typing;
touch footprint.in
Use DOCK6 to generate footprints
dock6 -i footprint.in
Use the following lines to answer the prompted questions.
conformer_search_type rigid use_internal_energy no ligand_atom_file 2nnq_lig_min.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename 2nnq_lig_with.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ../1.dockprep/2nnq_rec_withH.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 12 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix footprint.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Once everything is successful these output files should be generated. (footprint.out_footprint_scored.txt, footprint.out_hbond_scored.txt, footprint.out_scored.mol2)
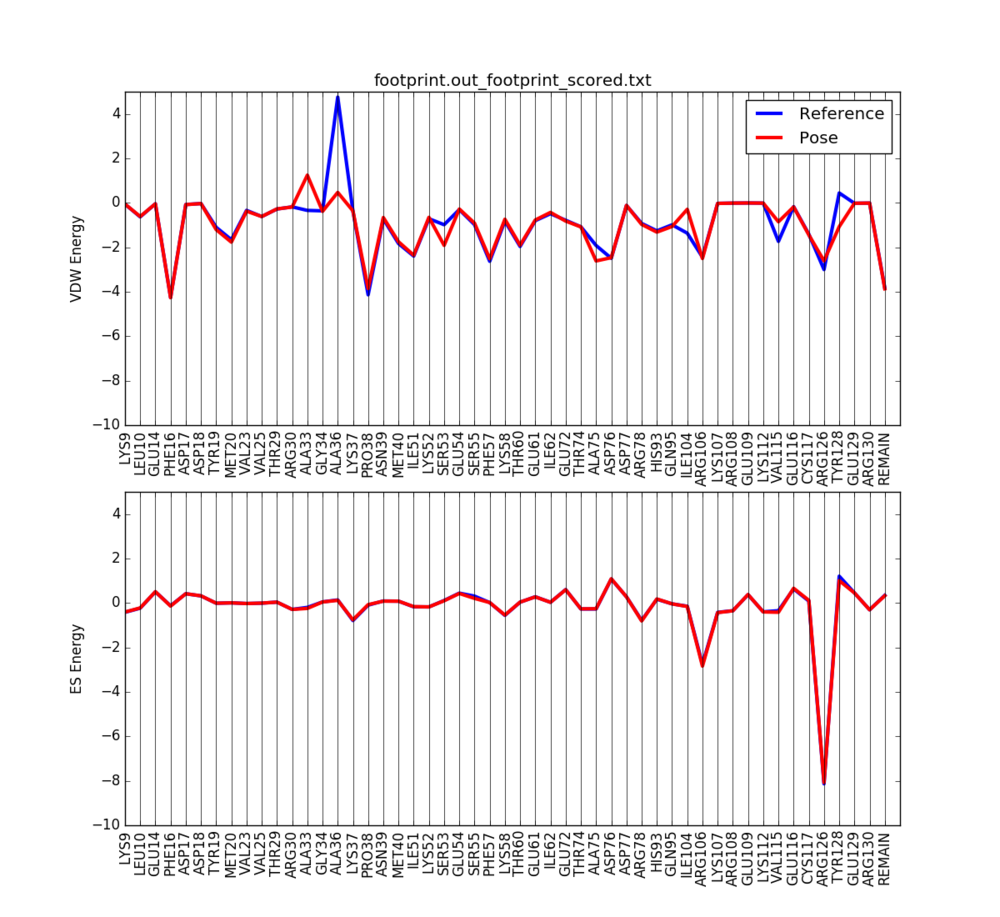
Use a python script to visualize the molecular footprint. The script can be accessed in the previous DOCK tutorials. Once the script is used, the molecular footprint should be similar to the image below. Notice the large deviations in energy at different amino acid residues. Those residues contribute more towards, the interaction between the ligand and therefore can be identified as important towards the binding of new ligands which can replace the original ligand in the PDB file.
VI. Virtual Screen
Virtual screening is the method of screening a ligand library (drug-like molecules) to filter the best ligands which can bind to the binding site of a specific receptor. Here we will be using a ligand library which contains 25000 molecules to select the best ligands which can be used to replace the original ligand that the PDB file contained.
Move to the directory 7.virtual_screen. Copy the ligand library to the same directory.
Create a new input file for virtual screen.
touch virtual.in
Use the input file to perform virtual screen using DOCK6.
dock6 -i virtual.in
Use the following lines to answer the prompted questions.
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file small_ligand_library.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../3.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix virtual.out write_orientations no num_scored_conformers 1 rank_ligands no
Since the ligand library contains 25000 molecules, it is going to take a long time to complete the virtual screen. Therefore we are going to use the mpi version of DOCK to complete the virtual screen. Therefore you can either terminate the virtual screen which is already running by pressing ctrl+c or let it run.
VII.Virtual Screen (MPI)
Until now we used the head node of the seawulf cluster. By using the mpi version of DOCK we will be using 4 processors that contain 28 nodes in each and it will complete the virtual screen quicker. Move to a new directory. (8.virtual_screen_mpi) Copy the input file and the ligand database file from 7.virtual_screen directory.
To submit the job to the seawulf cluster we are using a new file. (virtual.sh)
vim virtual.sh
Type the following lines in the new file.
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=4:ppn=28 #PBS -q long #PBS -N 2nnq.virtual #PBS -V cd $PBS_O_WORKDIR mpirun -np 112 dock6.mpi -i virtual.in -o 2nnq.virtual.mpi.out
The virtual screen job can be submitted to the cluster using the following command.
qsub virtual.sh
Type the following command to check the status of the job and other jobs that you submitted via your user login.
qstat -u username
VIII.Cartesian Minimization
Here we will use the docked molecules and perform a cartesian minimization of them.
Move to the directory 8.cartesianmin.
Create a new input file for the minimization.
touch min.in
Use the input file to perform the cartesian minimization using DOCK6.
dock6 -i min.in
Use the following lines to answer the prompted questions.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 2nnq.virtualscreen_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../1.dockprep/2nnq.rec.charged.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 2nnq.virtualscreen.minimized write_orientations no num_scored_conformers 1 rank_ligands no
Create a new submission script for the minimization.
touch min.sh
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=28 #PBS -q long #PBS -N cartesian min #PBS -V cd $PBS_O_WORKDIR dock6 -i min.in -o min.out
IX.Rescoring Docked Molecules
We would also like to rank our docked ligands and extract the 100 best ligands (which have the most negative, lowest scores). Footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score will all be used by Dock to rescore the virtual screen.
Move to the directory 9.rescore.
Create a new input file for the rescoring.
touch rescore.in
Use the input file to perform the cartesian minimization using DOCK6.
dock6 -i rescore.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../8.cartesianmin/2nnq.virtualscreen.minimized_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_energy no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_use_footprint_reference_mol2 yes descriptor_fps_footprint_reference_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 descriptor_fps_foot_compare_type Euclidean descriptor_fps_normalize_foot no descriptor_fps_foot_comp_all_residue yes descriptor_fps_receptor_filename ../1.dockprep/2nnq.rec.withH.charged.mol2 descriptor_fps_vdw_att_exp 6 descriptor_fps_vdw_rep_exp 12 descriptor_fps_vdw_rep_rad_scale 1 descriptor_fps_use_distance_dependent_dielectric yes descriptor_fps_dielectric 4.0 descriptor_fps_vdw_fp_scale 1 descriptor_fps_es_fp_scale 1 descriptor_fps_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../4.dock/2nnq.lig.min_scored.mol2 descriptor_hungarian_ref_filename ../4.dock/2nnq.lig.min_scored.mol2 descriptor_hungarian_matching_coeff -5 descriptor_hungarian_rmsd_coeff 1 descriptor_volume_reference_mol2_filename ../4.dock/2nnq.lig.min_scored.mol2 descriptor_volume_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hungarian 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no