Difference between revisions of "2019 Denovo design tutorial 1 with PDB 2BXF"
Stonybrook (talk | contribs) (Created page with "==De novo design with PDB 1C87 focused fragment library== ===Preparing The Files=== First of all, we need to create a directory for the fragment library. mkdir fraglib Mov...") |
Stonybrook (talk | contribs) (Undo revision 12724 by Stonybrook (talk)) |
||
| (23 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | == | + | =='''I.'' de-novo'' design with PDB 2BXF (Focused)'''== |
| − | === | + | ===Fragment Libraries=== |
| − | + | A focused fragment library will be used in this tutorial, in order to attempt building the same ligand. A focused fragment library can be generated using the same ligand. | |
| + | Create a new directory for the fragment library. | ||
mkdir fraglib | mkdir fraglib | ||
| − | + | Inside the fraglib directory create a new input file for fragment generation. | |
| + | touch fraglib.in | ||
| + | Generate the fragments by calling the input file through DOCK6 | ||
| + | dock6 -i fraglib.in | ||
| + | |||
| + | Answer the prompted questions interactively using the following lines. | ||
conformer_search_type flex | conformer_search_type flex | ||
write_fragment_libraries yes | write_fragment_libraries yes | ||
| Line 13: | Line 19: | ||
fragment_library_freq_cutoff 1 | fragment_library_freq_cutoff 1 | ||
fragment_library_sort_method freq | fragment_library_sort_method freq | ||
| − | fragment_library_trans_origin no | + | fragment_library_trans_origin no |
| − | use_internal_energy | + | use_internal_energy yes |
| − | ligand_atom_file / | + | internal_energy_rep_exp 12 |
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file /Path_to_file/2BXF_lig_withH.mol2 | ||
limit_max_ligands no | limit_max_ligands no | ||
skip_molecule no | skip_molecule no | ||
read_mol_solvation no | read_mol_solvation no | ||
| − | calculate_rmsd no | + | calculate_rmsd no |
use_database_filter no | use_database_filter no | ||
| − | orient_ligand no | + | orient_ligand yes |
| + | automated_matching yes | ||
| + | receptor_site_file /Path_to_file/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
bump_filter no | bump_filter no | ||
score_molecules no | score_molecules no | ||
| Line 28: | Line 42: | ||
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| − | ligand_outfile_prefix | + | ligand_outfile_prefix output |
write_orientations no | write_orientations no | ||
num_scored_conformers 1 | num_scored_conformers 1 | ||
rank_ligands no | rank_ligands no | ||
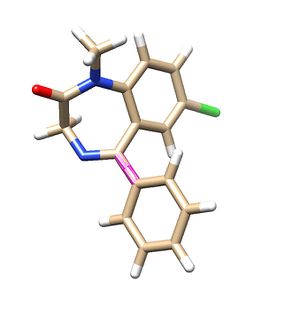
| − | fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, | + | Once fragment generation is completed, following files will be generated. (fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat) Open the mol2 files using chimera and check if the fragments match with the ligand used for fragment generation. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | conformer_search_type denovo | + | [[File:Sidechain.jpg|thumb|center|300px|2BXF sidechain fragment]] |
| − | dn_fraglib_scaffold_file fraglib_scaffold.mol2 | + | |
| − | dn_fraglib_linker_file fraglib_linker.mol2 | + | ===Focused Denovo Growth=== |
| − | dn_fraglib_sidechain_file fraglib_sidechain.mol2 | + | The generated fragments will be used to perform the de novo dock. Here the fragments will added together restricted by different properties like maximum layers, formal charge and molecular weight. Before each connection, DOCK will check with the torsion environment file created in frgment generation to see if that particular connection is seen before. If not the connection won't be made. |
| − | dn_user_specified_anchor | + | |
| − | + | Create a new directory for denovo dock. | |
| + | mkdir denovo | ||
| + | |||
| + | Create a new input file for denovo dock. | ||
| + | touch denovo.in | ||
| + | |||
| + | Run the input file through dock. | ||
| + | dock6 -i denovo.in | ||
| + | |||
| + | Answer the prompted question interactively using the following lines. | ||
| + | |||
| + | conformer_search_type denovo | ||
| + | conformer_search_type denovo | ||
| + | dn_fraglib_scaffold_file /8.fraglib/fraglib_scaffold.mol2 | ||
| + | dn_fraglib_linker_file /8.fraglib/fraglib_linker.mol2 | ||
| + | dn_fraglib_sidechain_file /8.fraglib/fraglib_sidechain.mol2 | ||
| + | dn_user_specified_anchor no | ||
dn_use_torenv_table yes | dn_use_torenv_table yes | ||
| − | dn_torenv_table fraglib_torenv.dat | + | dn_torenv_table /8.fraglib/fraglib_torenv.dat |
dn_sampling_method graph | dn_sampling_method graph | ||
dn_graph_max_picks 30 | dn_graph_max_picks 30 | ||
| Line 56: | Line 79: | ||
dn_graph_temperature 100.0 | dn_graph_temperature 100.0 | ||
dn_pruning_conformer_score_cutoff 100.0 | dn_pruning_conformer_score_cutoff 100.0 | ||
| − | dn_pruning_conformer_score_scaling_factor | + | dn_pruning_conformer_score_scaling_factor 1.0 |
dn_pruning_clustering_cutoff 100.0 | dn_pruning_clustering_cutoff 100.0 | ||
dn_constraint_mol_wt 550.0 | dn_constraint_mol_wt 550.0 | ||
| Line 63: | Line 86: | ||
dn_heur_unmatched_num 1 | dn_heur_unmatched_num 1 | ||
dn_heur_matched_rmsd 2.0 | dn_heur_matched_rmsd 2.0 | ||
| − | dn_unique_anchors | + | dn_unique_anchors 2 |
dn_max_grow_layers 9 | dn_max_grow_layers 9 | ||
dn_max_root_size 25 | dn_max_root_size 25 | ||
| Line 70: | Line 93: | ||
dn_max_scaffolds_per_layer 1 | dn_max_scaffolds_per_layer 1 | ||
dn_write_checkpoints yes | dn_write_checkpoints yes | ||
| − | dn_write_prune_dump | + | dn_write_prune_dump no |
dn_write_orients no | dn_write_orients no | ||
| − | dn_write_growth_trees | + | dn_write_growth_trees yes |
| − | dn_output_prefix | + | dn_output_prefix 2BXF.final |
use_internal_energy yes | use_internal_energy yes | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
| − | internal_energy_cutoff 100.0 | + | internal_energy_cutoff 100.0 |
use_database_filter no | use_database_filter no | ||
orient_ligand yes | orient_ligand yes | ||
automated_matching yes | automated_matching yes | ||
| − | receptor_site_file | + | receptor_site_file /2.surface_spheres/selected_spheres.sph |
max_orientations 1000 | max_orientations 1000 | ||
critical_points no | critical_points no | ||
| Line 94: | Line 117: | ||
grid_score_vdw_scale 1 | grid_score_vdw_scale 1 | ||
grid_score_es_scale 1 | grid_score_es_scale 1 | ||
| − | grid_score_grid_prefix | + | grid_score_grid_prefix /3.boxgrid/grid |
multigrid_score_secondary no | multigrid_score_secondary no | ||
dock3.5_score_secondary no | dock3.5_score_secondary no | ||
| Line 119: | Line 142: | ||
simplex_grow_tors_premin_iterations 0 | simplex_grow_tors_premin_iterations 0 | ||
simplex_random_seed 0 | simplex_random_seed 0 | ||
| − | simplex_restraint_min no | + | simplex_restraint_min no |
atom_model all | atom_model all | ||
vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| Line 125: | Line 148: | ||
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
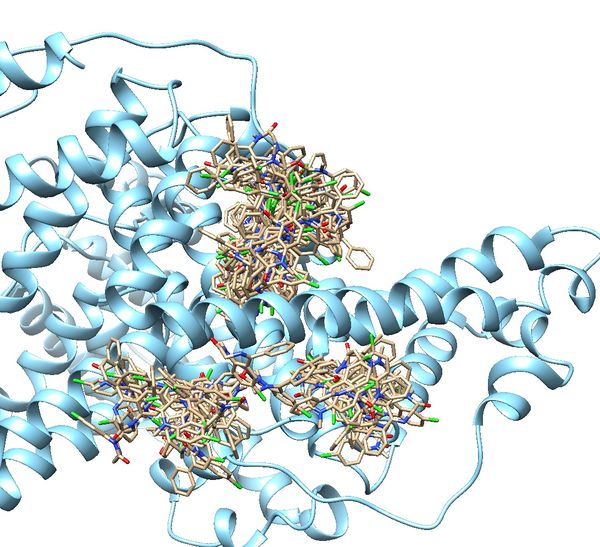
| − | + | [[File:Denovo rescored.jpg|thumb|center|600px|2BXF growthed molecules]] | |
| − | + | ||
| − | + | ===Focused Denovo Rescore=== | |
| − | + | ||
| − | + | Create a new directory | |
| − | + | mkdir focuseddenovorescore | |
| − | + | ||
| − | + | Copy your reference .mol2 file into this directory | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Create an input file | ||
| + | touch rescore.in | ||
| − | + | Complete the input file using the automated questions from dock | |
| − | + | dock6 -i rescore.in | |
| − | |||
| − | |||
| − | |||
conformer_search_type rigid | conformer_search_type rigid | ||
| Line 150: | Line 167: | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
internal_energy_cutoff 100.0 | internal_energy_cutoff 100.0 | ||
| − | ligand_atom_file | + | ligand_atom_file output.denovo_build.mol2 |
limit_max_ligands no | limit_max_ligands no | ||
skip_molecule no | skip_molecule no | ||
| − | read_mol_solvation no | + | read_mol_solvation no |
| − | calculate_rmsd | + | calculate_rmsd no |
| − | |||
| − | |||
use_database_filter no | use_database_filter no | ||
orient_ligand no | orient_ligand no | ||
| Line 180: | Line 195: | ||
descriptor_use_multigrid_score no | descriptor_use_multigrid_score no | ||
descriptor_use_continuous_score no | descriptor_use_continuous_score no | ||
| − | descriptor_use_footprint_similarity | + | descriptor_use_footprint_similarity yes |
| − | descriptor_use_pharmacophore_score | + | descriptor_use_pharmacophore_score yes |
descriptor_use_tanimoto yes | descriptor_use_tanimoto yes | ||
descriptor_use_hungarian yes | descriptor_use_hungarian yes | ||
| − | descriptor_use_volume_overlap no | + | descriptor_use_volume_overlap yes |
| − | + | descriptor_fps_score_use_footprint_reference_mol2 yes | |
| − | + | descriptor_fps_score_footprint_reference_mol2_filename candidate_mol_gen_frag.mol2 | |
| + | descriptor_fps_score_foot_compare_type Euclidean | ||
| + | descriptor_fps_score_normalize_foot no | ||
| + | descriptor_fps_score_foot_comp_all_residue yes | ||
| + | descriptor_fps_score_receptor_filename ../../dock/2BXF.rec.withH.charged.mol2 | ||
| + | descriptor_fps_score_vdw_att_exp 6 | ||
| + | descriptor_fps_score_vdw_rep_exp 12 | ||
| + | descriptor_fps_score_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_score_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_score_dielectric 4.0 | ||
| + | descriptor_fps_score_vdw_fp_scale 1 | ||
| + | descriptor_fps_score_es_fp_scale 1 | ||
| + | descriptor_fps_score_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename candidate_mol_gen_frag.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename candidate_mol_gen_frag.mol2 | ||
| + | descriptor_hms_score_ref_filename candidate_mol_gen_frag.mol2 | ||
descriptor_hms_score_matching_coeff -5 | descriptor_hms_score_matching_coeff -5 | ||
descriptor_hms_score_rmsd_coeff 1 | descriptor_hms_score_rmsd_coeff 1 | ||
| + | descriptor_volume_score_reference_mol2_filename candidate_mol_gen_frag.mol2 | ||
| + | descriptor_volume_score_overlap_compute_method analytical | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
descriptor_weight_fingerprint_tanimoto -1 | descriptor_weight_fingerprint_tanimoto -1 | ||
descriptor_weight_hms_score 1 | descriptor_weight_hms_score 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file ../../dock/box/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file ../../dock/dock/flex.defn | ||
| + | flex_drive_file ../../dock/dock/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn | ||
| + | pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | You will now have a file in your directory called descriptor.output_scored.mol2 which can be loaded into Chimera using ViewDock with your original ligand .mol2 file. | ||
| + | |||
| + | =='''II.''de-novo'' growth with PDB 2BXF (Generic)'''== | ||
| + | In this section, the de novo growth will be done using a fragment library generated by many random ligands. This allows DOCK to generate novel ligands which are not restricted by the fragments and torsion environment of the crystal ligand. | ||
| + | |||
| + | In the previous section, single grid was used as the scoring function to generate the ligands. In this section, the interactions of the ligand with the most significant residues will be considered for the scoring function. In order to do so, multigrid scoring method where each of the significant residues will be described using unique grids, and a common grid for the rest of the residues. | ||
| + | |||
| + | ===Specifying Primary Residues=== | ||
| + | First, the significant residues for the binding of the ligand has to be specified. Create unique directory for the generic de novo growth. Create an input file. (rescore.in) Use the following lines to generate the input file. | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy no | ||
| + | ligand_atom_file ../2BXF_lig_withH.mol2 (use the ligand mol2 file) | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary yes | ||
| + | footprint_similarity_score_secondary no | ||
| + | fps_score_use_footprint_reference_mol2 yes | ||
| + | fps_score_footprint_reference_mol2_filename ../2BXF_lig_withH.mol2 (use the ligand mol2 file) | ||
| + | fps_score_foot_compare_type Euclidean | ||
| + | fps_score_normalize_foot no | ||
| + | fps_score_foot_comp_all_residue no | ||
| + | fps_score_choose_foot_range_type threshold | ||
| + | fps_score_vdw_threshold 1 | ||
| + | fps_score_es_threshold 0.5 | ||
| + | fps_score_hb_threshold 0.5 | ||
| + | fps_score_use_remainder yes | ||
| + | fps_score_receptor_filename ../2BXF_rec.mol2 (use the mol2 file for the receptor) | ||
| + | fps_score_vdw_att_exp 6 | ||
| + | fps_score_vdw_rep_exp 12 | ||
| + | fps_score_vdw_rep_rad_scale 1 | ||
| + | fps_score_use_distance_dependent_dielectric yes | ||
| + | fps_score_dielectric 4.0 | ||
| + | fps_score_vdw_fp_scale 1 | ||
| + | fps_score_es_fp_scale 1 | ||
| + | fps_score_hb_fp_scale 0 | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
gbsa_zou_score_secondary no | gbsa_zou_score_secondary no | ||
gbsa_hawkins_score_secondary no | gbsa_hawkins_score_secondary no | ||
| Line 200: | Line 318: | ||
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| − | ligand_outfile_prefix | + | ligand_outfile_prefix rescore.out |
| + | write_footprints yes | ||
| + | write_hbonds no | ||
write_orientations no | write_orientations no | ||
num_scored_conformers 1 | num_scored_conformers 1 | ||
rank_ligands no | rank_ligands no | ||
| − | |||
| − | |||
| − | |||
| − | + | dock6 -i rescore.in -o 2BXF.footprint_rescore.out | |
| + | |||
| + | This will generate three output files. | ||
| + | 2BXF.footprint_rescore.out | ||
| + | rescore.out_footprint_scored.txt | ||
| + | rescore.out_scored.mol2 | ||
| + | |||
| + | Use these files to specify the primary residues. Create a new file specify.sh using the following lines which are taken from previous tutorials and altered accordingly. | ||
| + | |||
| + | #!/bin/bash | ||
| + | grep -A 1 "range_union" 2BXF.footprint_rescore.out | | ||
| + | grep -v "range_union" | | ||
| + | grep -v "\-" | | ||
| + | sed -e '{s/,/\n/g}' | | ||
| + | sed -e '{s/ //g}' | | ||
| + | sed '/^$/d' | | ||
| + | sort -n | | ||
| + | uniq > temp.dat | ||
| + | for i in `cat temp.dat`; do printf "%0*d\n" 3 $i; done > 2BXF.primary_residues.dat | ||
| + | for RES in `cat temp.dat` | ||
| + | do | ||
| + | grep " ${RES} " rescore.out_footprint_scored.txt | | ||
| + | awk -v temp=${RES} '{if ($2 == temp) print $0;}' | | ||
| + | awk '{print $1 " " $3 " " $4}' >> reference.txt | ||
| + | done | ||
| + | grep "remainder" rescore.out_footprint_scored.txt | | ||
| + | sed -e '{s/,/ /g}' | | ||
| + | tr -d '\n' | | ||
| + | awk '{print $2 " " $3 " " $6}' >> reference.txt | ||
| + | mv reference.txt 2BXF.reference.txt | ||
| + | rm temp.dat | ||
| + | |||
| + | Use either of the following commands to specify the primary residues using the above file. | ||
| + | chmod 770 specify.sh | ||
| + | ./specify.sh | ||
| + | or | ||
| + | bash specify.sh | ||
| + | |||
| + | This will generate another two files which specify the primary residues of the binding site of your receptor. | ||
| + | 2BXF.primary_residues.dat | ||
| + | 2BXF.reference.txt | ||
| + | If it is successful, 13 residues should be specified as primary residues. (this is just for this system and the number of primary residues can change from system to system) | ||
| + | |||
| + | In the next section, grids will be generated for the primary residues and a common grid for the rest of the residues. | ||
| + | |||
| + | ===Generation of grids=== | ||
| + | Create a new directory (multigrid) | ||
| + | This will be done by a script file and two input files. The structure of each file is given below. | ||
| + | |||
| + | First input file (2BXF.multigrid.in) | ||
| + | |||
| + | compute_grids yes | ||
| + | grid_spacing 0.4 | ||
| + | output_molecule yes | ||
| + | contact_score no | ||
| + | chemical_score no | ||
| + | energy_score yes | ||
| + | energy_cutoff_distance 9999 | ||
| + | atom_model a | ||
| + | attractive_exponent 6 | ||
| + | repulsive_exponent 9 | ||
| + | distance_dielectric yes | ||
| + | dielectric_factor 4 | ||
| + | bump_filter yes | ||
| + | bump_overlap 0.75 | ||
| + | receptor_file temp.mol2 | ||
| + | box_file ../../../dock/3.boxgrid/2BXF.box.pdb (use the virtual box generated in dock previously) | ||
| + | vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | chemical_definition_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn | ||
| + | score_grid_prefix temp.rec | ||
| + | receptor_out_file temp.rec.grid.mol2 | ||
| + | |||
| + | Second input file (2BXF.reference_multigridmin.in) | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../../2BXF_lig_withH.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary yes | ||
| + | multigrid_score_secondary no | ||
| + | multigrid_score_rep_rad_scale 1.0 | ||
| + | multigrid_score_vdw_scale 1.0 | ||
| + | multigrid_score_es_scale 1.0 | ||
| + | multigrid_score_number_of_grids 14 (Should be the total number of primary residues + 1) | ||
| + | multigrid_score_grid_prefix0 2BXF.resid_380 | ||
| + | multigrid_score_grid_prefix1 2BXF.resid_383 | ||
| + | multigrid_score_grid_prefix2 2BXF.resid_384 | ||
| + | multigrid_score_grid_prefix3 2BXF.resid_387 | ||
| + | multigrid_score_grid_prefix4 2BXF.resid_399 | ||
| + | multigrid_score_grid_prefix5 2BXF.resid_403 | ||
| + | multigrid_score_grid_prefix6 2BXF.resid_407 | ||
| + | multigrid_score_grid_prefix7 2BXF.resid_426 | ||
| + | multigrid_score_grid_prefix8 2BXF.resid_429 | ||
| + | multigrid_score_grid_prefix9 2BXF.resid_445 | ||
| + | multigrid_score_grid_prefix10 2BXF.resid_446 | ||
| + | multigrid_score_grid_prefix11 2BXF.resid_449 | ||
| + | multigrid_score_grid_prefix12 2BXF.resid_481 | ||
| + | multigrid_score_grid_prefix13 2BXF.resid_remaining (rest of the residues) | ||
| + | multigrid_score_fp_ref_mol no | ||
| + | multigrid_score_fp_ref_text yes | ||
| + | multigrid_score_footprint_text ../2BXF.reference.txt (generated earlier when specifying the primary residues) | ||
| + | multigrid_score_foot_compare_type Euclidean | ||
| + | multigrid_score_normalize_foot no | ||
| + | multigrid_score_vdw_euc_scale 1.0 | ||
| + | multigrid_score_es_euc_scale 1.0 | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix output | ||
| + | write_footprints no | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Use the following script to generate the grids. (multigrid.sh) | ||
| − | == | + | export PRIMARY_RES=` cat ../2BXF.primary_residues.dat | sed -e 's/\n/ /g' ` |
| − | + | export DOCKHOME="/gpfs/projects/AMS536/zzz.programs/dock6" | |
| + | python ${DOCKHOME}/bin/multigrid_fp_gen.py ../../2BXF_rec.mol2 2BXF.resid 2BXF.multigrid.in ${PRIMARY_RES} | ||
| + | rm temp.mol2 | ||
| + | rm 2BXF.resid_*.rec.grid.mol2 | ||
| + | ${DOCKHOME}/bin/dock6 -i 2BXF.reference_multigridmin.in -o 2BXF.reference_multigridmin.out | ||
| + | mv output_scored.mol2 2BXF.lig.multigridmin.mol2 | ||
| + | cp 2BXF.lig.multigridmin.mol2 ../multi-grid | ||
| − | + | Run the above script to generate the grids | |
| + | bash multigrid.sh | ||
| − | + | If it is successful, grids will be generated. Check 2BXF.lig.multigridmin.out file to make sure everything is okay. | |
| − | + | ===Running de novo growth === | |
| + | In this section, a de novo growth will be done using the grids generated above and a provided fragment library where the fragments are generated from random drug like ligandsprovided in the dock6 distribution in the parameters directory. | ||
| − | + | Go one directory back from multigrid and create a new directory, denovo. Copy the neccesary fragment library files needed into this directory. (scaffold,linker,sidechain,torsion env ..etc) Then create an input file for denovo growth using the lines below and save as denovo.in. | |
conformer_search_type denovo | conformer_search_type denovo | ||
| − | dn_fraglib_scaffold_file | + | dn_fraglib_scaffold_file fraglib_scaffold.mol2 |
| − | dn_fraglib_linker_file | + | dn_fraglib_linker_file fraglib_linker.mol2 |
| − | dn_fraglib_sidechain_file | + | dn_fraglib_sidechain_file fraglib_sidechain.mol2 |
dn_user_specified_anchor no | dn_user_specified_anchor no | ||
dn_use_torenv_table yes | dn_use_torenv_table yes | ||
| − | dn_torenv_table | + | dn_torenv_table fraglib_torenv.dat |
dn_sampling_method graph | dn_sampling_method graph | ||
dn_graph_max_picks 30 | dn_graph_max_picks 30 | ||
| Line 236: | Line 509: | ||
dn_pruning_conformer_score_scaling_factor 1.0 | dn_pruning_conformer_score_scaling_factor 1.0 | ||
dn_pruning_clustering_cutoff 100.0 | dn_pruning_clustering_cutoff 100.0 | ||
| − | dn_constraint_mol_wt | + | dn_constraint_mol_wt 550.0 |
dn_constraint_rot_bon 15 | dn_constraint_rot_bon 15 | ||
dn_constraint_formal_charge 2.0 | dn_constraint_formal_charge 2.0 | ||
| Line 242: | Line 515: | ||
dn_heur_matched_rmsd 2.0 | dn_heur_matched_rmsd 2.0 | ||
dn_unique_anchors 1 | dn_unique_anchors 1 | ||
| − | dn_max_grow_layers | + | dn_max_grow_layers 9 |
dn_max_root_size 25 | dn_max_root_size 25 | ||
dn_max_layer_size 25 | dn_max_layer_size 25 | ||
dn_max_current_aps 5 | dn_max_current_aps 5 | ||
| − | dn_max_scaffolds_per_layer | + | dn_max_scaffolds_per_layer 1 |
dn_write_checkpoints yes | dn_write_checkpoints yes | ||
dn_write_prune_dump no | dn_write_prune_dump no | ||
dn_write_orients no | dn_write_orients no | ||
dn_write_growth_trees no | dn_write_growth_trees no | ||
| − | dn_output_prefix | + | dn_output_prefix output |
use_internal_energy yes | use_internal_energy yes | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
| Line 258: | Line 531: | ||
orient_ligand yes | orient_ligand yes | ||
automated_matching yes | automated_matching yes | ||
| − | receptor_site_file ../ | + | receptor_site_file ../2.surface_spheres/selected_spheres.sph |
max_orientations 1000 | max_orientations 1000 | ||
critical_points no | critical_points no | ||
| Line 290: | Line 563: | ||
descriptor_multigrid_score_vdw_scale 1.0 | descriptor_multigrid_score_vdw_scale 1.0 | ||
descriptor_multigrid_score_es_scale 1.0 | descriptor_multigrid_score_es_scale 1.0 | ||
| − | descriptor_multigrid_score_number_of_grids | + | descriptor_multigrid_score_number_of_grids 14 |
| − | descriptor_multigrid_score_grid_prefix0 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix0 ../15.multi-grid/2BXF.resid_380 |
| − | descriptor_multigrid_score_grid_prefix1 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix1 ../15.multi-grid/2BXF.resid_383 |
| − | descriptor_multigrid_score_grid_prefix2 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix2 ../15.multi-grid/2BXF.resid_384 |
| − | descriptor_multigrid_score_grid_prefix3 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix3 ../15.multi-grid/2BXF.resid_387 |
| − | descriptor_multigrid_score_grid_prefix4 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix4 ../15.multi-grid/2BXF.resid_399 |
| − | descriptor_multigrid_score_grid_prefix5 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix5 ../15.multi-grid/2BXF.resid_403 |
| − | descriptor_multigrid_score_grid_prefix6 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix6 ../15.multi-grid/2BXF.resid_407 |
| − | descriptor_multigrid_score_grid_prefix7 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix7 ../15.multi-grid/2BXF.resid_426 |
| − | descriptor_multigrid_score_grid_prefix8 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix8 ../15.multi-grid/2BXF.resid_429 |
| − | descriptor_multigrid_score_grid_prefix9 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix9 ../15.multi-grid/2BXF.resid_445 |
| − | descriptor_multigrid_score_grid_prefix10 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix10 ../15.multi-grid/2BXF.resid_446 |
| − | descriptor_multigrid_score_grid_prefix11 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix11 ../15.multi-grid/2BXF.resid_449 |
| − | descriptor_multigrid_score_grid_prefix12 ../multi-grid/ | + | descriptor_multigrid_score_grid_prefix12 ../15.multi-grid/2BXF.resid_481 |
| − | descriptor_multigrid_score_grid_prefix13 ../ | + | descriptor_multigrid_score_grid_prefix13 ../15.multi-grid/2BXF.resid_remaining |
| − | + | descriptor_multigrid_score_fp_ref_mol yes | |
| − | + | descriptor_multigrid_score_footprint_ref ../4.dock/2BXF.lig.min_scored.mol2 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | descriptor_multigrid_score_fp_ref_mol | ||
| − | |||
| − | |||
descriptor_multigrid_score_foot_compare_type Euclidean | descriptor_multigrid_score_foot_compare_type Euclidean | ||
descriptor_multigrid_score_normalize_foot no | descriptor_multigrid_score_normalize_foot no | ||
| Line 345: | Line 605: | ||
simplex_restraint_min no | simplex_restraint_min no | ||
atom_model all | atom_model all | ||
| − | vdw_defn_file . | + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn |
| − | flex_defn_file . | + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn |
| − | flex_drive_file . | + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl |
| + | |||
| + | Use the following script (denovo.sh) to submit the job to the cluster. | ||
| + | #!/bin/bash | ||
| + | #PBS -l walltime=48:00:00 | ||
| + | #PBS -l nodes=1:ppn=28 | ||
| + | #PBS -q long | ||
| + | #PBS -N 2BXF_denovo | ||
| + | #PBS -V | ||
| + | cd $PBS_O_WORKDIR | ||
| + | dock6 -i denovo.in -o 2BXF_denovo.out | ||
| + | |||
| + | Use the following command to submit the job. | ||
| + | qsub denovo.sh | ||
Latest revision as of 13:58, 10 March 2020
Contents
I. de-novo design with PDB 2BXF (Focused)
Fragment Libraries
A focused fragment library will be used in this tutorial, in order to attempt building the same ligand. A focused fragment library can be generated using the same ligand.
Create a new directory for the fragment library.
mkdir fraglib
Inside the fraglib directory create a new input file for fragment generation.
touch fraglib.in
Generate the fragments by calling the input file through DOCK6
dock6 -i fraglib.in
Answer the prompted questions interactively using the following lines.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file /Path_to_file/2BXF_lig_withH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file /Path_to_file/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix output write_orientations no num_scored_conformers 1 rank_ligands no
Once fragment generation is completed, following files will be generated. (fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat) Open the mol2 files using chimera and check if the fragments match with the ligand used for fragment generation.
Focused Denovo Growth
The generated fragments will be used to perform the de novo dock. Here the fragments will added together restricted by different properties like maximum layers, formal charge and molecular weight. Before each connection, DOCK will check with the torsion environment file created in frgment generation to see if that particular connection is seen before. If not the connection won't be made.
Create a new directory for denovo dock.
mkdir denovo
Create a new input file for denovo dock.
touch denovo.in
Run the input file through dock.
dock6 -i denovo.in
Answer the prompted question interactively using the following lines.
conformer_search_type denovo
conformer_search_type denovo dn_fraglib_scaffold_file /8.fraglib/fraglib_scaffold.mol2 dn_fraglib_linker_file /8.fraglib/fraglib_linker.mol2 dn_fraglib_sidechain_file /8.fraglib/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table /8.fraglib/fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 2 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees yes dn_output_prefix 2BXF.final use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file /2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix /3.boxgrid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Focused Denovo Rescore
Create a new directory
mkdir focuseddenovorescore
Copy your reference .mol2 file into this directory
Create an input file
touch rescore.in
Complete the input file using the automated questions from dock
dock6 -i rescore.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file output.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename candidate_mol_gen_frag.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../../dock/2BXF.rec.withH.charged.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename candidate_mol_gen_frag.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename candidate_mol_gen_frag.mol2 descriptor_hms_score_ref_filename candidate_mol_gen_frag.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename candidate_mol_gen_frag.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file ../../dock/box/vdw_AMBER_parm99.defn flex_defn_file ../../dock/dock/flex.defn flex_drive_file ../../dock/dock/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
You will now have a file in your directory called descriptor.output_scored.mol2 which can be loaded into Chimera using ViewDock with your original ligand .mol2 file.
II.de-novo growth with PDB 2BXF (Generic)
In this section, the de novo growth will be done using a fragment library generated by many random ligands. This allows DOCK to generate novel ligands which are not restricted by the fragments and torsion environment of the crystal ligand.
In the previous section, single grid was used as the scoring function to generate the ligands. In this section, the interactions of the ligand with the most significant residues will be considered for the scoring function. In order to do so, multigrid scoring method where each of the significant residues will be described using unique grids, and a common grid for the rest of the residues.
Specifying Primary Residues
First, the significant residues for the binding of the ligand has to be specified. Create unique directory for the generic de novo growth. Create an input file. (rescore.in) Use the following lines to generate the input file.
conformer_search_type rigid use_internal_energy no ligand_atom_file ../2BXF_lig_withH.mol2 (use the ligand mol2 file) limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ../2BXF_lig_withH.mol2 (use the ligand mol2 file) fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue no fps_score_choose_foot_range_type threshold fps_score_vdw_threshold 1 fps_score_es_threshold 0.5 fps_score_hb_threshold 0.5 fps_score_use_remainder yes fps_score_receptor_filename ../2BXF_rec.mol2 (use the mol2 file for the receptor) fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 12 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix rescore.out write_footprints yes write_hbonds no write_orientations no num_scored_conformers 1 rank_ligands no
dock6 -i rescore.in -o 2BXF.footprint_rescore.out
This will generate three output files.
2BXF.footprint_rescore.out rescore.out_footprint_scored.txt rescore.out_scored.mol2
Use these files to specify the primary residues. Create a new file specify.sh using the following lines which are taken from previous tutorials and altered accordingly.
#!/bin/bash
grep -A 1 "range_union" 2BXF.footprint_rescore.out |
grep -v "range_union" |
grep -v "\-" |
sed -e '{s/,/\n/g}' |
sed -e '{s/ //g}' |
sed '/^$/d' |
sort -n |
uniq > temp.dat
for i in `cat temp.dat`; do printf "%0*d\n" 3 $i; done > 2BXF.primary_residues.dat
for RES in `cat temp.dat`
do
grep " ${RES} " rescore.out_footprint_scored.txt |
awk -v temp=${RES} '{if ($2 == temp) print $0;}' |
awk '{print $1 " " $3 " " $4}' >> reference.txt
done
grep "remainder" rescore.out_footprint_scored.txt |
sed -e '{s/,/ /g}' |
tr -d '\n' |
awk '{print $2 " " $3 " " $6}' >> reference.txt
mv reference.txt 2BXF.reference.txt
rm temp.dat
Use either of the following commands to specify the primary residues using the above file.
chmod 770 specify.sh ./specify.sh
or
bash specify.sh
This will generate another two files which specify the primary residues of the binding site of your receptor.
2BXF.primary_residues.dat 2BXF.reference.txt
If it is successful, 13 residues should be specified as primary residues. (this is just for this system and the number of primary residues can change from system to system)
In the next section, grids will be generated for the primary residues and a common grid for the rest of the residues.
Generation of grids
Create a new directory (multigrid) This will be done by a script file and two input files. The structure of each file is given below.
First input file (2BXF.multigrid.in)
compute_grids yes grid_spacing 0.4 output_molecule yes contact_score no chemical_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file temp.mol2 box_file ../../../dock/3.boxgrid/2BXF.box.pdb (use the virtual box generated in dock previously) vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn chemical_definition_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn score_grid_prefix temp.rec receptor_out_file temp.rec.grid.mol2
Second input file (2BXF.reference_multigridmin.in)
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../../2BXF_lig_withH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary yes multigrid_score_secondary no multigrid_score_rep_rad_scale 1.0 multigrid_score_vdw_scale 1.0 multigrid_score_es_scale 1.0 multigrid_score_number_of_grids 14 (Should be the total number of primary residues + 1) multigrid_score_grid_prefix0 2BXF.resid_380 multigrid_score_grid_prefix1 2BXF.resid_383 multigrid_score_grid_prefix2 2BXF.resid_384 multigrid_score_grid_prefix3 2BXF.resid_387 multigrid_score_grid_prefix4 2BXF.resid_399 multigrid_score_grid_prefix5 2BXF.resid_403 multigrid_score_grid_prefix6 2BXF.resid_407 multigrid_score_grid_prefix7 2BXF.resid_426 multigrid_score_grid_prefix8 2BXF.resid_429 multigrid_score_grid_prefix9 2BXF.resid_445 multigrid_score_grid_prefix10 2BXF.resid_446 multigrid_score_grid_prefix11 2BXF.resid_449 multigrid_score_grid_prefix12 2BXF.resid_481 multigrid_score_grid_prefix13 2BXF.resid_remaining (rest of the residues) multigrid_score_fp_ref_mol no multigrid_score_fp_ref_text yes multigrid_score_footprint_text ../2BXF.reference.txt (generated earlier when specifying the primary residues) multigrid_score_foot_compare_type Euclidean multigrid_score_normalize_foot no multigrid_score_vdw_euc_scale 1.0 multigrid_score_es_euc_scale 1.0 dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix output write_footprints no write_orientations no num_scored_conformers 1 rank_ligands no
Use the following script to generate the grids. (multigrid.sh)
export PRIMARY_RES=` cat ../2BXF.primary_residues.dat | sed -e 's/\n/ /g' `
export DOCKHOME="/gpfs/projects/AMS536/zzz.programs/dock6"
python ${DOCKHOME}/bin/multigrid_fp_gen.py ../../2BXF_rec.mol2 2BXF.resid 2BXF.multigrid.in ${PRIMARY_RES}
rm temp.mol2
rm 2BXF.resid_*.rec.grid.mol2
${DOCKHOME}/bin/dock6 -i 2BXF.reference_multigridmin.in -o 2BXF.reference_multigridmin.out
mv output_scored.mol2 2BXF.lig.multigridmin.mol2
cp 2BXF.lig.multigridmin.mol2 ../multi-grid
Run the above script to generate the grids
bash multigrid.sh
If it is successful, grids will be generated. Check 2BXF.lig.multigridmin.out file to make sure everything is okay.
Running de novo growth
In this section, a de novo growth will be done using the grids generated above and a provided fragment library where the fragments are generated from random drug like ligandsprovided in the dock6 distribution in the parameters directory.
Go one directory back from multigrid and create a new directory, denovo. Copy the neccesary fragment library files needed into this directory. (scaffold,linker,sidechain,torsion env ..etc) Then create an input file for denovo growth using the lines below and save as denovo.in.
conformer_search_type denovo dn_fraglib_scaffold_file fraglib_scaffold.mol2 dn_fraglib_linker_file fraglib_linker.mol2 dn_fraglib_sidechain_file fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix output use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../2.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score yes descriptor_use_pharmacophore_score no descriptor_use_tanimoto no descriptor_use_hungarian no descriptor_use_volume_overlap no descriptor_multigrid_score_rep_rad_scale 1.0 descriptor_multigrid_score_vdw_scale 1.0 descriptor_multigrid_score_es_scale 1.0 descriptor_multigrid_score_number_of_grids 14 descriptor_multigrid_score_grid_prefix0 ../15.multi-grid/2BXF.resid_380 descriptor_multigrid_score_grid_prefix1 ../15.multi-grid/2BXF.resid_383 descriptor_multigrid_score_grid_prefix2 ../15.multi-grid/2BXF.resid_384 descriptor_multigrid_score_grid_prefix3 ../15.multi-grid/2BXF.resid_387 descriptor_multigrid_score_grid_prefix4 ../15.multi-grid/2BXF.resid_399 descriptor_multigrid_score_grid_prefix5 ../15.multi-grid/2BXF.resid_403 descriptor_multigrid_score_grid_prefix6 ../15.multi-grid/2BXF.resid_407 descriptor_multigrid_score_grid_prefix7 ../15.multi-grid/2BXF.resid_426 descriptor_multigrid_score_grid_prefix8 ../15.multi-grid/2BXF.resid_429 descriptor_multigrid_score_grid_prefix9 ../15.multi-grid/2BXF.resid_445 descriptor_multigrid_score_grid_prefix10 ../15.multi-grid/2BXF.resid_446 descriptor_multigrid_score_grid_prefix11 ../15.multi-grid/2BXF.resid_449 descriptor_multigrid_score_grid_prefix12 ../15.multi-grid/2BXF.resid_481 descriptor_multigrid_score_grid_prefix13 ../15.multi-grid/2BXF.resid_remaining descriptor_multigrid_score_fp_ref_mol yes descriptor_multigrid_score_footprint_ref ../4.dock/2BXF.lig.min_scored.mol2 descriptor_multigrid_score_foot_compare_type Euclidean descriptor_multigrid_score_normalize_foot no descriptor_multigrid_score_vdw_euc_scale 1.0 descriptor_multigrid_score_es_euc_scale 1.0 descriptor_weight_multigrid_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Use the following script (denovo.sh) to submit the job to the cluster.
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=28 #PBS -q long #PBS -N 2BXF_denovo #PBS -V cd $PBS_O_WORKDIR dock6 -i denovo.in -o 2BXF_denovo.out
Use the following command to submit the job.
qsub denovo.sh