Difference between revisions of "2023 DOCK tutorial 2 with PDBID 3WZE"
Stonybrook (talk | contribs) (→Surface Generation) |
BrockBoysan (talk | contribs) (→Grid generation) |
||
| (37 intermediate revisions by one other user not shown) | |||
| Line 110: | Line 110: | ||
cp -r directory_1 directory_2 | cp -r directory_1 directory_2 | ||
| + | |||
| + | To move a directory from your local machine to Seawulf, from your local directory, type: | ||
| + | |||
| + | scp -r 'NETID@login.seawulf.stonybrook.edu:/FULL PATH' . | ||
| + | |||
| + | To move a file from your local machine to Seawulf, from your local directory, type: | ||
| + | |||
| + | scp FILENAME NETID@login.seawulf.stonybrook.edu:/FULL PATH | ||
== '''Directory Organization''' == | == '''Directory Organization''' == | ||
| Line 131: | Line 139: | ||
Open 3WZE in Chimera or ChimeraX to examine it. It should contain the structures of VEGFR II (henceforth called the "receptor"), the bound inhibitor sorafinib (a ligand), and a number of crystallographic waters. | Open 3WZE in Chimera or ChimeraX to examine it. It should contain the structures of VEGFR II (henceforth called the "receptor"), the bound inhibitor sorafinib (a ligand), and a number of crystallographic waters. | ||
| + | |||
| + | == Streamlined Prep == | ||
| + | |||
| + | While following each of the sections below can be informative, I've also included this streamlined prep section to save you from having to do some extra steps. This abbreviated protocol works for 3WZE, but it may not work correctly for your protein, so be sure to check things like charges, hydrogens, presence of non-standard residues, and missing loops before applying this process to your protein. | ||
| + | |||
| + | '''ChimeraX''': | ||
| + | |||
| + | 1. Download 3WZE.pdb from the Protein Data Bank, and open it in ChimeraX | ||
| + | |||
| + | 2. Follow the steps in the Filling in Missing Loops section. You can skip this if your protein doesn't have any missing loops. | ||
| + | |||
| + | 3. Type "delete ligand" | ||
| + | |||
| + | 4. Type "dockprep" | ||
| + | |||
| + | 5. Save the receptor as a .mol2 file. | ||
== Removing Solvent == | == Removing Solvent == | ||
| Line 147: | Line 171: | ||
'''ChimeraX''': | '''ChimeraX''': | ||
| − | 1. Type "delete solvent" into the command line. | + | 1. Type "show solvent" into the command line. This isn't necessary, but it allows you to see whether step 2 worked. The waters should appear as little red balls: |
| + | |||
| + | [[File: DougVSwater.png|thumb|center|1000px]] | ||
| + | |||
| + | 2. Type "delete solvent" into the command line. | ||
| + | |||
| + | [[File: DougVSnowater.png|thumb|center|1000px]] | ||
== Removing the Ligand and Non-Standard Residues == | == Removing the Ligand and Non-Standard Residues == | ||
To perform a virtual screen, the binding site of the receptor must be empty of any bound ligand. 3ZWE has the inhibitor sorafinib bound within the active site. Both Chimera and ChimeraX designate it as a ligand, and as a non-standard residue. If you look at the list of non-standard residues, you'll notice that 3WZE also contains a free-floating acetone molecule and a cysteine residue which has formed a disulfide bond with the reducing agent dithiothreitol. Neither the acetone molecule, nor the DTT-cysteine are reflective of the receptor's state in nature, and are instead artifacts of the crystallization process used to solve the receptor's structure. Therefore, they should be removed alongside the sorafinib ligand. | To perform a virtual screen, the binding site of the receptor must be empty of any bound ligand. 3ZWE has the inhibitor sorafinib bound within the active site. Both Chimera and ChimeraX designate it as a ligand, and as a non-standard residue. If you look at the list of non-standard residues, you'll notice that 3WZE also contains a free-floating acetone molecule and a cysteine residue which has formed a disulfide bond with the reducing agent dithiothreitol. Neither the acetone molecule, nor the DTT-cysteine are reflective of the receptor's state in nature, and are instead artifacts of the crystallization process used to solve the receptor's structure. Therefore, they should be removed alongside the sorafinib ligand. | ||
| + | |||
| + | [[File: DougVScysdtt.png|thumb|center|1000px]] | ||
| + | Here's a view of 682 which displays the abnormal disulfide bond with DTT. | ||
| + | |||
| + | [[File: DougVSacetone.png|thumb|center|1000px]] | ||
| + | And this is the acetone molecule that we should get rid of. | ||
'''Chimera''': | '''Chimera''': | ||
| Line 163: | Line 199: | ||
1. Type "delete ligand" (the acetone, sorafinib, and cysteine-DTT are all designated as ligands, so this one command will delete them all) | 1. Type "delete ligand" (the acetone, sorafinib, and cysteine-DTT are all designated as ligands, so this one command will delete them all) | ||
| − | Check that the cysteine sidechain is intact after the deletion of its bonded DTT. In the case of 3WZE, the cysteine sidechain remains intact after running the above commands in either program. | + | Check that the cysteine sidechain is intact after the deletion of its bonded DTT. In the case of 3WZE, the cysteine sidechain remains intact after running the above commands in either program: |
| + | |||
| + | [[File: DougVScysnoDTT.png|thumb|center|1000px]] | ||
== Filling in Missing Loops == | == Filling in Missing Loops == | ||
| Line 234: | Line 272: | ||
'''ChimeraX''': | '''ChimeraX''': | ||
| − | 1. Type “addh” into the command line. | + | 1. Type “addh” into the command line. |
| + | |||
| + | 2. Type "show sidechain" into the command line. The molecule should now look liike the image below, where the white atoms are hydrogens: | ||
| + | |||
| + | [[File: DougVShyd.png|thumb|center|1000px]] | ||
This will populate the entire protein with hydrogens, but it will also make the N and C termini charged when we add charges, which isn’t reflective of the charge state of those residues in nature. The receptor in 3WZE is only a portion of the VEGFR II protein, so the backbone ends in 3WZE aren't the protein's true N and C termini. Chimera and ChimeraX has no way of knowing this however, so it will add one too many hydrogens at the N terminal side of the receptor and one too few at the C terminal side of the receptor. This incorrect hydrogenation may not be reflective of the protein in nature, but it is reflective of the protein construct as it was crystallized, so it doesn’t need to be corrected as long as the charged termini aren’t close enough to the active site to mess with docking. In the case of 3WZE, they are not too close, so we can leave the construct's ends as they are. | This will populate the entire protein with hydrogens, but it will also make the N and C termini charged when we add charges, which isn’t reflective of the charge state of those residues in nature. The receptor in 3WZE is only a portion of the VEGFR II protein, so the backbone ends in 3WZE aren't the protein's true N and C termini. Chimera and ChimeraX has no way of knowing this however, so it will add one too many hydrogens at the N terminal side of the receptor and one too few at the C terminal side of the receptor. This incorrect hydrogenation may not be reflective of the protein in nature, but it is reflective of the protein construct as it was crystallized, so it doesn’t need to be corrected as long as the charged termini aren’t close enough to the active site to mess with docking. In the case of 3WZE, they are not too close, so we can leave the construct's ends as they are. | ||
| Line 245: | Line 287: | ||
1. Use Tools->Structure Editing->Add Charge | 1. Use Tools->Structure Editing->Add Charge | ||
| + | |||
| + | Standard residues: AMBER ff14SB | ||
| + | Other residues: AM1-BCC | ||
2. Click OK in the pop-up window | 2. Click OK in the pop-up window | ||
| Line 251: | Line 296: | ||
1. Type "addcharge" | 1. Type "addcharge" | ||
| + | 2. Save the molecule as a mol2 file | ||
| + | 3. Open the mol2 file in a text editor in order to check that charges were added. Atomic charges will appear in the last column of the mol2 file, as highlighted below: | ||
| + | |||
| + | [[File: Mthd.PNG|thumb|center|1000px]] | ||
= '''Preparing the Ligand''' = | = '''Preparing the Ligand''' = | ||
| + | |||
| + | '''Chimera''': | ||
| + | |||
| + | 1. Use Select->Residue->BAX to select the ligand. | ||
| + | |||
| + | 2. Use Select->Invert (all models) | ||
| + | |||
| + | 3. Use Actions->Atoms/Bonds->Delete | ||
| + | |||
| + | 4. Use Tools->Structure Editing->AddH | ||
| + | |||
| + | 5. Return to the paper, and check whether the hydrogens added by ChimeraX is accurate to the actual chemistry. Be considerate of any diagrams depicting hbonds or charge-charge interactions when looking for clues. Also be mindful of pH of the solution used to generate crystals. In the case of 3WZE, the hydrogens added by Addh are correct. | ||
| + | |||
| + | 6. Save the ligand as a mol2 file. | ||
| + | |||
| + | 7. Open it in a text viewer such as Text Editor, or open it from the command line with commands like "vi" or "nano" | ||
| + | |||
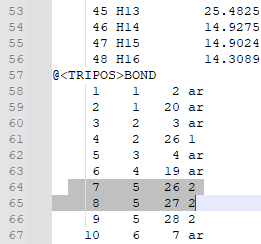
| + | 8. Check that the bonds are correct. For this particular ligand, bonds 7 and 8 were erroneously labeled as double bonds when they should be amide bonds, and the corresponding Nitrogen atoms (N12 and N14, or atoms 26 and 27 in the mol2 file) are labeled as “N.pl3” instead of “N.am”. | ||
| + | |||
| + | 9. Change the “2” in the last column of bond 7’s and bond 8’s rows to “am”. This will change the double bond to an amide bond. | ||
| + | Change the “N.pl3” to “N.am” in the rows for atoms 26 and 27. This will make Nitrogen 12 and 14 into amide nitrogens as they should be. | ||
| + | |||
| + | 11. Save the new mol2 file | ||
| + | |||
| + | 12. Open the mol2 in Chimera | ||
| + | |||
| + | 13. Use Tools->Structure Editing->Add Charge to add charges | ||
| + | |||
| + | Standard residues: AMBER ff14SB | ||
| + | Other residues: AM1-BCC | ||
| + | |||
| + | 14. Click OK in the pop-up window. For 3WZE, an additional pop-up will appear asking you to set the overall charge of the molecule because it isn't a standard molecule that Chimera would know the charge of. Set the charge based on your knowledge of chemistry and the information in the paper. For 3WZE, set it to 0. | ||
| + | |||
| + | 15. Save the molecule as a final mol2 file. | ||
| + | |||
| + | |||
| + | '''ChimeraX''': | ||
| + | |||
| + | 1. Open the original 3WZE.pdb file in ChimeraX | ||
| + | |||
| + | 2. Select the Sorafinib inhibitor by typing "select :BAX". A green outline should appear around the ligand. The non-standard residue name assigned to sorafinib is "BAX" for some reason. | ||
| + | |||
| + | [[File: DougVSlig1.png|thumb|center|1000px]] | ||
| + | |||
| + | 3. Use Select->Invert to select everything except the ligand. Everything except the ligand should now have a green outline. | ||
| + | |||
| + | [[File: DougVSlig2.png|thumb|center|1000px]] | ||
| + | |||
| + | 4. Type "delete sel". You should just be left with your ligand: | ||
| + | |||
| + | [[File: DougVSlig3.png|thumb|center|1000px]] | ||
| + | |||
| + | 5. Type "addh" | ||
| + | |||
| + | [[File: DougVSlig4.png|thumb|center|1000px]] | ||
| + | |||
| + | 6. Return to the paper, and check whether the hydrogens added by ChimeraX is accurate to the actual chemistry. Be considerate of any diagrams depicting hbonds or charge-charge interactions when looking for clues. Also be mindful of pH of the solution used to generate crystals. In the case of 3WZE, the hydrogens added by Addh are correct. | ||
| + | |||
| + | 7. Save the ligand as a mol2 file. | ||
| + | |||
| + | 8. Open it in a text viewer such as Text Editor, or open it from the command line with commands like "vi" or "nano" | ||
| + | |||
| + | 9. Check that the bonds are correct. For this particular ligand, bonds 7 and 8 were erroneously labeled as double bonds when they should be amide bonds, and the corresponding Nitrogen atoms (N12 and N14, or atoms 26 and 27 in the mol2 file) are labeled as “N.pl3” instead of “N.am”. | ||
| + | |||
| + | 10. Change the “2” in the last column of bond 7’s and bond 8’s rows (shown below) to “am”. This will change the double bond to an amide bond. | ||
| + | |||
| + | [[File: DougVSlig5.PNG|thumb|center|1000px]] | ||
| + | |||
| + | Change the “N.pl3” to “N.am” in the rows for atoms 26 and 27. This will make Nitrogen 12 and 14 into amide nitrogens as they should be. | ||
| + | |||
| + | [[File: DougVSlig6.PNG|thumb|center|1000px]] | ||
| + | |||
| + | 11. Save the new mol2 file | ||
| + | |||
| + | 12. Open the mol2 in ChimeraX | ||
| + | |||
| + | 13. With most ligands, you should be able to simply type “addcharge” to assign charges to each atom and save as a mol2 file. However, doing that with this molecule will erroneously assign a +1 charge somewhere and yield an error message. To avoid that, force ChimeraX to assign a neutral charge by typing “addcharge nonstd :BAX BAX 0” | ||
| + | |||
| + | 14. Save the molecule as a final mol2 file. | ||
= ''' Surface & Spheres '''= | = ''' Surface & Spheres '''= | ||
| + | |||
| + | At this stage, move directories to Seawulf. | ||
| + | |||
==='''Surface Generation'''=== | ==='''Surface Generation'''=== | ||
| − | The generation of the surface of the receptor protein will have better visualization of the binding site of the receptor. In order to do so, load the mol2 file of 3WZE receptor in Chimera: | + | The generation of the surface of the receptor protein will have better visualization of the binding site of the receptor. In order to do so, load the mol2 file of 3WZE receptor without hydrogens in Chimera: |
Action < Surface < show | Action < Surface < show | ||
| + | Then you will be able to visualize the surface of your protein in Chimera, it will be like something in the image below: | ||
| + | [[File:3WZEsurface.png|center|600px]] | ||
Then we will save the dms(molecular surface) file as 3WZE_rec.dms, which will be used in the next step of spheres generation: | Then we will save the dms(molecular surface) file as 3WZE_rec.dms, which will be used in the next step of spheres generation: | ||
| Line 274: | Line 407: | ||
First create a new file “INSPH” by typing the following into terminal: | First create a new file “INSPH” by typing the following into terminal: | ||
vi INSPH | vi INSPH | ||
| − | + | Note that the name "INSPH" is not arbitrary. Attempting to use a differently named input file will yield an error message. | |
| + | |||
| + | Type the following information into this file (anything after # are comments that are references for reading only, and should not be putting into the file) : | ||
3WZE_rec.dms #Molecular Surface file | 3WZE_rec.dms #Molecular Surface file | ||
R #This line specifies whether to make the spheres in the model’s exterior (R) or interior (L) | R #This line specifies whether to make the spheres in the model’s exterior (R) or interior (L) | ||
| Line 282: | Line 417: | ||
1.4 #Minimum radius of each sphere | 1.4 #Minimum radius of each sphere | ||
3WZE.sph #Name of the output file that will be made | 3WZE.sph #Name of the output file that will be made | ||
| − | Then save the file and run the following command on terminal: note that if you want to re-run this command, make sure you remove file “OUTSPH” before your re-run because this file cannot be overwritten. | + | Then save the file and run the following command on terminal: note that if you want to re-run this command, make sure you remove file “OUTSPH” before your re-run because this file cannot be overwritten. Also remove any temp files that sphgen made. Also bear in mind that sphgen will crash if your protein is too big (greater than ~900 amino acids), so if you're getting an error message, try deleting part of your protein which isn't needed for docking before generating the surface and trying to generate spheres. |
sphgen -i INSPH -o OUTSPH | sphgen -i INSPH -o OUTSPH | ||
The output file “3WZE.sph” can be visualized by using Chimera, load the output file along with the receptor protein mol2 file, the two structures should be overlapping: | The output file “3WZE.sph” can be visualized by using Chimera, load the output file along with the receptor protein mol2 file, the two structures should be overlapping: | ||
| + | [[File:3WZEsphere.png|center|600px]] | ||
==='''Spheres selection'''=== | ==='''Spheres selection'''=== | ||
| Line 290: | Line 426: | ||
sphere_selector 3WZE.sph 3WZE_lig.mol2 10.0 | sphere_selector 3WZE.sph 3WZE_lig.mol2 10.0 | ||
#sphere_selector [sphere output file] [ligand mol2 file] [radius for distance from ligand] | #sphere_selector [sphere output file] [ligand mol2 file] [radius for distance from ligand] | ||
| + | Open this file in Chimera along with the receptor file, you will see something like this: | ||
| + | [[File:3WZEselectedsph.png|center|600px]] | ||
= '''Box and Grid '''= | = '''Box and Grid '''= | ||
| Line 299: | Line 437: | ||
vi showbox.in | vi showbox.in | ||
Then type the following information into the file (anything after # are comments that are references for reading only, and should not be putting into the file) : | Then type the following information into the file (anything after # are comments that are references for reading only, and should not be putting into the file) : | ||
| − | + | Y #Indicates that we want to make a box | |
| − | + | 8.0 #Extra length beyond the spheres to be included by the box in all directions | |
| − | + | ../002_surface_spheres/selected_spheres.sph #Indicates that the box should be constructed to encompass the spheres in this file. | |
| − | + | 1 #This specifies which cluster of spheres should be used to generate the box. | |
| − | + | 3WZE.box.pdb #Name of output file | |
| + | |||
Then, run the following code to get the boxed structure: | Then, run the following code to get the boxed structure: | ||
showbox < showbox.in | showbox < showbox.in | ||
| Line 311: | Line 450: | ||
vi grid.in | vi grid.in | ||
Then type the following information into the file: | Then type the following information into the file: | ||
| + | allow_non_integral_charges no | ||
compute_grids yes | compute_grids yes | ||
grid_spacing 0.4 | grid_spacing 0.4 | ||
| Line 322: | Line 462: | ||
distance_dielectric yes | distance_dielectric yes | ||
dielectric_factor 4 | dielectric_factor 4 | ||
| + | allow_non_integral_charges yes | ||
bump_filter yes | bump_filter yes | ||
bump_overlap 0.75 | bump_overlap 0.75 | ||
| Line 331: | Line 472: | ||
grid -i grid.in -o gridinfo.out | grid -i grid.in -o gridinfo.out | ||
This will generate three files: “gridinfo.out”, “grid.bmp”, and “grid.nrg”. Open “gridinfo.out” and check if the information about the receptor matches the that from the paper and your previous preparations. Also check to make sure there are no error messages at the bottom. | This will generate three files: “gridinfo.out”, “grid.bmp”, and “grid.nrg”. Open “gridinfo.out” and check if the information about the receptor matches the that from the paper and your previous preparations. Also check to make sure there are no error messages at the bottom. | ||
| + | |||
| + | Open "grid.nrg" and receptor file in Chimera, we will be able to see the energy grid of the protein: | ||
| + | [[File:3WZEgridenergy.png|center|600px]] | ||
| + | |||
| + | Open "grid.bmp" and receptor file in Chimera, we will be able to see the bump grid of the protein: | ||
| + | [[File:3WZEgridbump.png|center|600px]] | ||
='''Energy minimization '''= | ='''Energy minimization '''= | ||
Change the current working directory in terminal to do preparation for DOCK: | Change the current working directory in terminal to do preparation for DOCK: | ||
cd 004_dock | cd 004_dock | ||
| + | You may want to move over grid.nrg and grid.bmp as well. | ||
| + | |||
To improve the accuracy of DOCK calculation of ligand protein binding, we will have to minimize the potential energy of the ligand. To do so, create a new file “min.in” by using the following command: | To improve the accuracy of DOCK calculation of ligand protein binding, we will have to minimize the potential energy of the ligand. To do so, create a new file “min.in” by using the following command: | ||
vi min.in | vi min.in | ||
| Line 342: | Line 491: | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
internal_energy_cutoff 100.0 | internal_energy_cutoff 100.0 | ||
| − | ligand_atom_file ../000_files/ | + | ligand_atom_file ../000_files/3WZE_lig_wH.mol2 |
limit_max_ligands no | limit_max_ligands no | ||
skip_molecule no | skip_molecule no | ||
| Line 348: | Line 497: | ||
calculate_rmsd yes | calculate_rmsd yes | ||
use_rmsd_reference_mol yes | use_rmsd_reference_mol yes | ||
| − | rmsd_reference_filename ../000_files/ | + | rmsd_reference_filename ../000_files/3WZE_lig_wH.mol2 |
use_database_filter no | use_database_filter no | ||
orient_ligand no | orient_ligand no | ||
| Line 392: | Line 541: | ||
rank_ligands no | rank_ligands no | ||
Then run the following command through the use of DOCK: | Then run the following command through the use of DOCK: | ||
| − | dock6 -i min.in | + | dock6 -i min.in -o min.out |
| − | The output file “3WZE.lig.min_scored.mol2” is the potential energy minimized version of the ligand. | + | The output file “3WZE.lig.min_scored.mol2”(tan) is the potential energy minimized version of the ligand. Load this file along with ligand mol2 file(blue) in Chimera, you can compare the two molecules: |
| + | [[File:3WZEenergymin.png|center|600px]] | ||
= '''Reproducing the PDB's binding with DOCK''' = | = '''Reproducing the PDB's binding with DOCK''' = | ||
| Line 634: | Line 784: | ||
Copy the following into your file: | Copy the following into your file: | ||
| − | conformer_search_type | + | conformer_search_type rigid |
use_internal_energy no | use_internal_energy no | ||
ligand_atom_file 3WZE.lig.min_scored.mol2 | ligand_atom_file 3WZE.lig.min_scored.mol2 | ||
Latest revision as of 16:19, 12 February 2024
In this tutorial, you will learn how use the program DOCK6.10 to perform a virtual screen, in which you assess how well the molecules in a library of drug-like molecules bind to a protein of known structure.
Contents
Introduction
A protein whose function is found to be involved in one or more diseases may become a target for pharmaceutical design. Oftentimes, these pharmaceuticals are designed to compete with the enzyme's native substrate for the enzyme's active site, making many pharmaceutical molecules competitive inhibitors of their protein targets. If the target protein's structure is known, and the active site can be identified, then performing a virtual screen can be a monetarily and temporally efficient method of identifying molecules which are likely to bind well to the target's active site.
A virtual screen is set up by first preparing the enzyme's structure and the structure of its native substrate for docking, then the residues important for the native ligand to bind are identified by generating a footprint. A large library of drug-like molecules is then downloaded from a database such as ZINC (https://zinc.docking.org/), and, using the footprint and enzyme structure, docked into the enzyme using a program such as DOCK6.10. Results are then assessed to see which drug-like compounds match the native substrate's footprint profile and which are energetically comfortable within the simulated active site. Such molecules could then be tested biochemically for their ability to inhibit the target protein, sparing biochemists the hassle of having to test hundreds of thousands of compounds in physical screening experiments.
Software and Files
PDB 3WZE
PDB stands for "Protein Data Bank", which is a repository for the experimentally solved structures of proteins. Each protein structure is assigned a 4 digit code, and 3WZE is the code assigned to the solved structure of vascular endothelial growth factor receptor 2 (VEGFR2) bound to the inhibitor sorafenib. VEGFR2 is a receptor tyrosine kinase, meaning that it is an integral membrane protein that has an exofacial receptor domain, transmembrane domain, and cytofacial kinase domain. Because of the difficulties of protein purification, crystallization, and structure solution, many protein structures in the protein data bank are incomplete: lacking regions that are intrinsically disordered or otherwise not conducive to crystallization. PDB 3WZE is one such structure, because it only contains structural data for the cytofacial kinase domain of VEGFR2.
The active site of kinases is well characterized, and sorafenib is shown bound within it in PDB 3WZE, which will be useful for conducting later steps in the virtual screen.
Download the .pdb file of 3WZE, and use a program such as Chimera or ChimeraX to open and view it.
The Protein Data Bank home page can be found here:
DOCK6.10
DOCK is a program used to model conformations of molecules, and is typically used to find low potential energy conformations of drug-like molecules within the active sites of protein structures. Because molecules in nature tend to adopt conformations of the lowest possible potential energy, identifying low-energy conformations of molecules within proteins can be a useful way of identifying how such molecules may bind in nature. Thus, instead of having to manually test out libraries of thousands of molecules to identify potential therapeutics, a library of drug-like molecules can be docked into a protein of interest, vastly expediting the medicine development process at one of its earliest steps.
DOCK can be used for more purposes than drug identification though, and is capable of rigid and flexible docking, de novo design, and genetic algorithms. Rigid docking is when DOCK identifies low-potential energy conformations of a ligand within the target protein, but while keeping the ligand rigid. Flexible docking on the other hand allows for rotations around the ligand's internal rotatable bonds. De novo design is when DOCK attempts to build a new ligand outwards from a starting anchor specified by the user. Finally, genetic algorithms generate new ligands similarly tpo de novo design, but it also utilizes the principles of Darwinian evolution and natural selection to select for effective ligands over the course of multiple generations of ligand development.
For the purposes of this tutorial, you will only be using the rigid and flexible docking capabilities of DOCK. For more information on the other uses of DOCK, check the other tutorials in this wiki or consult the DOCK 6.10 User Manual:
https://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm#RigidandFlexibleLigandDocking
Chimera
Chimera is a visualization program that can display the 3-dimensional structures of molecules. Although Chimera is no longer actively developed, it remains a useful tool and an essential part of this tutorial. As you follow this tutorial, molecular visualization programs like Chimera and ChimeraX will be useful for preparing the system for the virtual screen and checking the results of each step. The home page of Chimera can be found here:
https://www.cgl.ucsf.edu/chimera/
ChimeraX (optional)
Chimera is now no longer actively developed, and has been succeeded by ChimeraX, which is developed by the same group. Although ChimeraX has lost some of the functionality of its predecessor, it has new capabilities to compensate, and it is easier to operate using typed commands, whereas Chimera requires clicking through menus. That being said, Chimera is still required for this tutorial because ChimeraX cannot open .sph files and it cannot save a surface as a .dms file.
Separate instructions for completing the tutorial with ChimeraX will be provided alongside the instructions for using Chimera in each section.
The home page for ChimeraX can be found here:
https://www.cgl.ucsf.edu/chimerax/
Alphafold (optional)
Alphafold is a protein structure prediction program from Google's Deepmind, and it was the first program to predict the structures of proteins in the annual CASP competition to within 90% accuracy in 2020 (https://predictioncenter.org/casp14/zscores_final.cgi). Using this program, one can generate a reasonably accurate prediction of a protein's structure using only its amino acid sequence. In the context of virtual screening, this means that a protein's structure no longer needs to be solved experimentally before one can embark on a virtual screen of the target. As long as the active site can be identified (which is often done by comparing the predicted structure to homologous proteins with solved structures), one can perform a virtual screen of a protein of unsolved structure.
Even without a university server, Alphafold can be used from within ChimeraX by going to Tools -> Structure Prediction -> Alphafold. This will bring up a menu in which you can paste an amino acid sequence for prediction, searching, or retrieval from the Alphafold database. All human proteins have already been predicted by Alphafold, and their structures can be easily retrieved using the protein's UniProt identifier and the Fetch button. Non-human proteins will have to be predicted from scratch by inputting their amino acid sequence and using the Predict button.
Because this tutorial will use PDB file 3WZE, which contains the solved structure of the Vascular Endothelial Growth Factor Receptor and a bound inhibitor called sorafenib, Alphafold will be unnecessary for this tutorial, but the broadened scope of what virtual screens are possible as a result of this program is worth noting nonetheless.
The Alphafold home page can be found here:
Using the Terminal
Seawulf uses the Linux language for various commands. Here are some of the basic commands that will make running the tutorial run smoothly.
The first command is cd, which is used to change into different directories. To go to a desired directory do:
cd /desired/directory
To move to a previous directory you were just in, type:
cd ../
The cd command can also be used to get into a folder, type:
cd /desired/folder
The next command is the ls command. This will list out all the files or folders in the directory you are currently in, type:
ls
To make a new directory, type:
mdkir name_of_directory
To remove a file, type:
rm file
To delete a directory and all its contents, type:
rm -r directory
To create a file in the terminal, type:
vi filename
This will open up a blank screen in which you can start typing. To edit the file, first type “i”. This will make the environment go to “INSERT” mode. You can now freely type. In order to save whatever you wrote in the file, click the escape button. This will remove you from “INSERT” mode. Next type:
:wq
This will save any changes you have made to your file
To move a file to a new location, type:
mv file /new/destination
To make a copy of a file, type:
cp file file_new_name
This command can also be used to copy directories as well, type:
cp -r directory_1 directory_2
To move a directory from your local machine to Seawulf, from your local directory, type:
scp -r 'NETID@login.seawulf.stonybrook.edu:/FULL PATH' .
To move a file from your local machine to Seawulf, from your local directory, type:
scp FILENAME NETID@login.seawulf.stonybrook.edu:/FULL PATH
Directory Organization
Having defined folders established before starting is a great way to maintain organization and clarity as you go.
000_files 001_structure 002_surface_spheres 003_gridbox 004_dock 005_virtual_screen 006_virtual_screen_mpi 007_cartesian_min 008_rescore
Additionally, file names should be clear and logical. We recommend starting with the PDB code followed by the receptor or ligand and ending with what changes were made. For example, 3WZE_lig_AddH_addCharge.mol2 indicates that this file is the ligand for 3WZE and hydrogens and charge have been added. Also, bear in mind that one should use underscores instead of spaces when naming files and directories. Otherwise, spaces in file or directory names can confuse many linux commands and make them mistake the names for command arguments.
Preparing the Receptor
Put the PDB file of 3WZE in files folder on Seawulf. You'll be doing initial receptor preparation steps on your local machine, but it's good to have the original file saved somewhere as a reference.
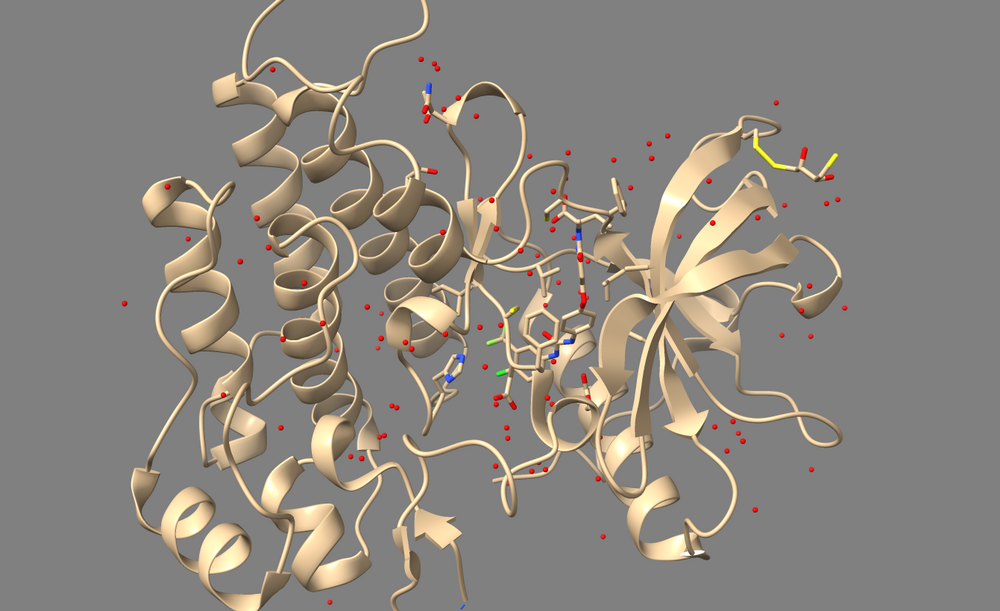
Open 3WZE in Chimera or ChimeraX to examine it. It should contain the structures of VEGFR II (henceforth called the "receptor"), the bound inhibitor sorafinib (a ligand), and a number of crystallographic waters.
Streamlined Prep
While following each of the sections below can be informative, I've also included this streamlined prep section to save you from having to do some extra steps. This abbreviated protocol works for 3WZE, but it may not work correctly for your protein, so be sure to check things like charges, hydrogens, presence of non-standard residues, and missing loops before applying this process to your protein.
ChimeraX:
1. Download 3WZE.pdb from the Protein Data Bank, and open it in ChimeraX
2. Follow the steps in the Filling in Missing Loops section. You can skip this if your protein doesn't have any missing loops.
3. Type "delete ligand"
4. Type "dockprep"
5. Save the receptor as a .mol2 file.
Removing Solvent
In Chimera, use Select->Residue->HOH to select water, then use Actions->Atoms/Bonds->Sphere to make the waters spheres, and finally use Actions->Atoms/Bonds->Show to reveal the waters. If you are using ChimeraX, simply type "show solvent" into the command line.
For the process of docking, these crystallographic waters within and around the receptor should be removed so that they don't impede the ability of ligands being docked to sample conformations on or within the receptor. However, if your receptor contains any water molecules which are essential for the structure or mechanism of it's active site, then those waters should be kept. Thus, one should read the paper associated with a PDB file to determine which waters, if any, should not be deleted.
In the case of 3WZE, there are no structurally important waters, so we'll delete all of them for the purposes of docking:
Chimera:
1. Use Select->Residue->HOH to select all water molecules.
2. Use Actions->Atoms/Bonds->delete to delete the selected molecules.
ChimeraX:
1. Type "show solvent" into the command line. This isn't necessary, but it allows you to see whether step 2 worked. The waters should appear as little red balls:
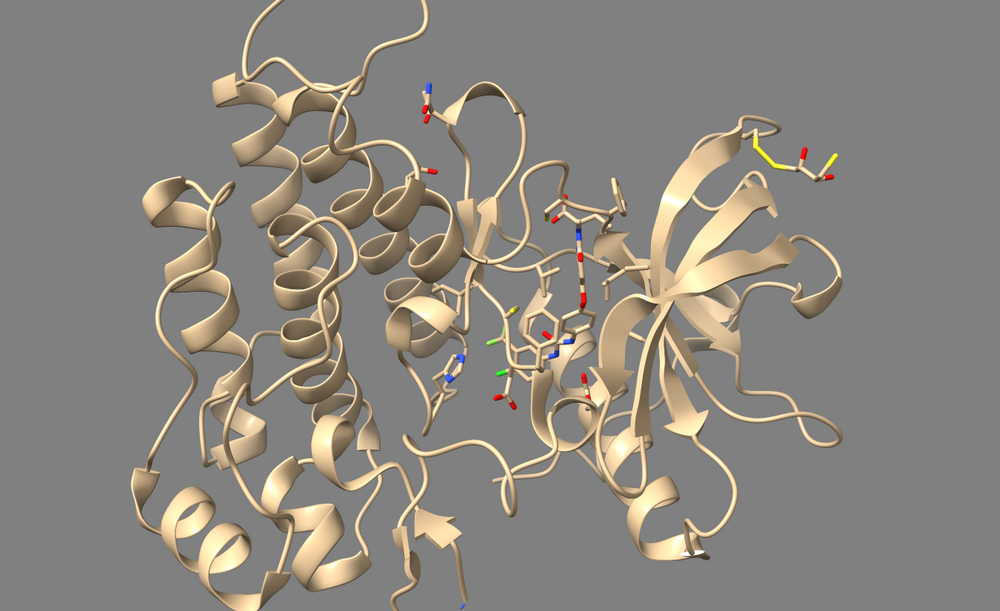
2. Type "delete solvent" into the command line.
Removing the Ligand and Non-Standard Residues
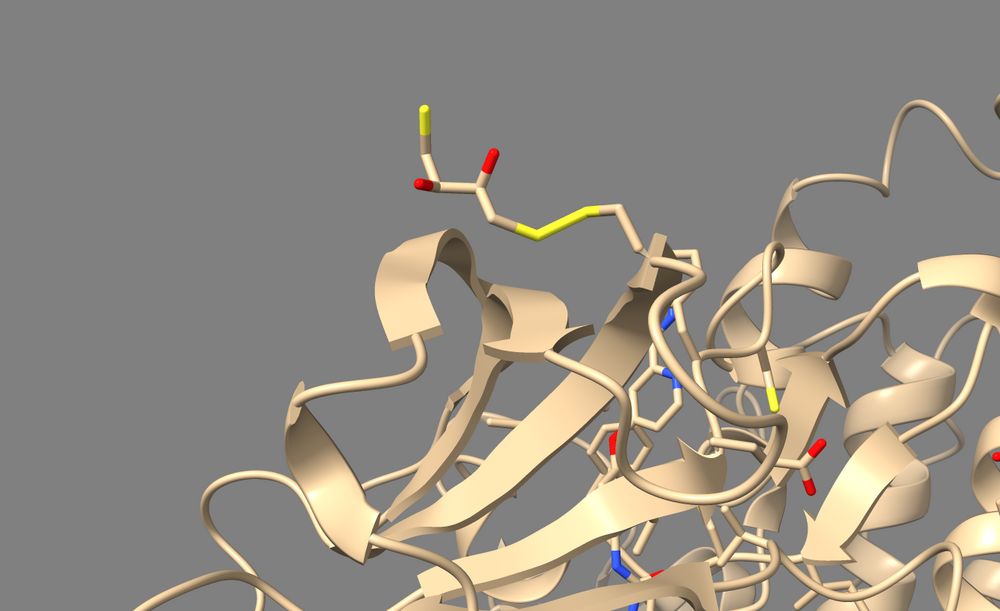
To perform a virtual screen, the binding site of the receptor must be empty of any bound ligand. 3ZWE has the inhibitor sorafinib bound within the active site. Both Chimera and ChimeraX designate it as a ligand, and as a non-standard residue. If you look at the list of non-standard residues, you'll notice that 3WZE also contains a free-floating acetone molecule and a cysteine residue which has formed a disulfide bond with the reducing agent dithiothreitol. Neither the acetone molecule, nor the DTT-cysteine are reflective of the receptor's state in nature, and are instead artifacts of the crystallization process used to solve the receptor's structure. Therefore, they should be removed alongside the sorafinib ligand.
Here's a view of 682 which displays the abnormal disulfide bond with DTT.
And this is the acetone molecule that we should get rid of.
Chimera:
1. Use Select->Residue->All non-standard (the acetone, sorafinib, and cysteine-DTT are all designated as non-standard residues)
2. Use Actions->Atoms/Bonds->Delete
ChimeraX:
1. Type "delete ligand" (the acetone, sorafinib, and cysteine-DTT are all designated as ligands, so this one command will delete them all)
Check that the cysteine sidechain is intact after the deletion of its bonded DTT. In the case of 3WZE, the cysteine sidechain remains intact after running the above commands in either program:
Filling in Missing Loops
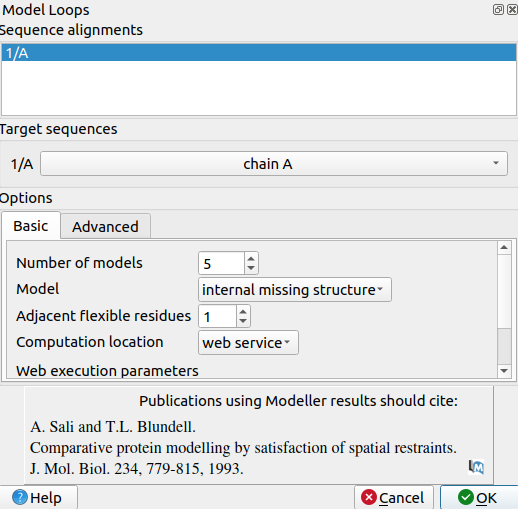
While examining the receptor in 3WZE, you may have noticed that there are some gaps in the protein's backbone which are bridged by dashed lines. These are gaps in the protein's structure, and are an artifact of the crystallization processes that are necessary for solving a structure. X-ray crystallography, the process by which many of the solved protein structures on the PDB are determined, can only be used to solve the structure of a protein that is arranged in a repeating, identical pattern (a crystal), so if any portions of the protein are intrinsically disordered, they can be too flexible to be consistent within the crystal, and may prevent crystallization entirely. As a result, it is very common for such disordered loops to be truncated prior to the protein purification and crystallization process.
While removing these loops are necessary for obtaining a structure, they can present problems for docking, especially if they would present charged terminal ends which could affect the docking process. In the case of 3WZE, the missing loops are too far from the active site to present a threat to docking, but it's good practice to fill them in anyway.
Chimera:
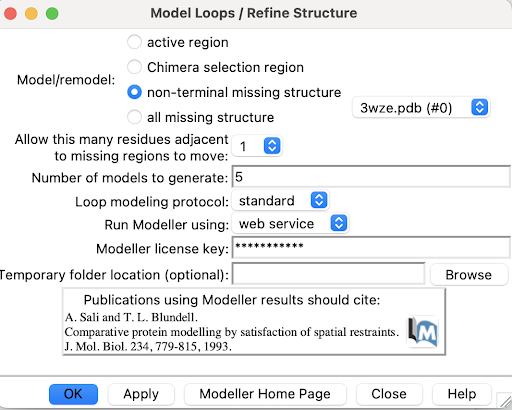
1. Use Tools->Structure Editing->Model/Refine Loops
A pop-up window will appear:
2. Select non-terminal missing structure. The only missing loops are internal ones, so the other options aren't relevant.
3. For "Allow this many adjacent to missing regions to move", set it to 1 or 0. Allowing some flexibility can help modeled loops look more organic, so 1 may be better than 0, but going any more residues than 1 risks altering the receptor's structure too much.
4. For "Number of models to generate", set it to 5.
5. Input a modeller license key. You can get one for free at https://salilab.org/modeller/registration.html
6. Click OK. After a few minutes, a window will pop-up with the Z scores for each of the 5 new chains. See which chain has the lowest Z score.
7. Select the chain with the lowest Z score using Select->Chains->A->name of chain with lowest Z score
8. Use Select->Invert (selected models) to select all chains except for the one with the lowest Z score
9. Use Actions->Atoms/Bonds-> delete to delete the other chains. Only your chain with filled in loops and lowest Z score will be left.
ChimeraX
1. Use Tools->Sequence->Show Sequence Viewer to open the Sequence Viewer
2. Within the Sequence Viewer pop-up window, click on Chain A and click show. A sidebar with the amino acid sequence of the receptor should appear
3. Use Tools->Structure Prediction->Model Loops to open a pop-up window for modeling missing loops
4. Set number of models to 5
5. Set Model to Internal missing structure. The only missing loops are internal ones, so the other options aren't relevant.
6. Set adjacent flexible residues to 1 or 0. Allowing some flexibility can help modeled loops look more organic, so 1 may be better than 0, but going any more residues than 1 risks altering the receptor's structure too much.
7. Input a modeller license key. You can get one for free at https://salilab.org/modeller/registration.html
8. Click OK. After a few minutes, a window will pop-up with the Z scores for each of the 5 new chains. See which chain has the lowest Z score.
9. Select the chain with the lowest Z score by typing "select #chainnmbr". For instance, my lowest scored chain was called #3.1, so I would type "select #3.1"
10. Use Select->Invert to invert your selection
11. Type "delete sel" to delete the selected chains. This will only leave your chain with the lowest Z score and filled loops.
Although you may see some differences in secondary structure (such as where beta sheets begin and end) in areas outside of just the disordered loops, by comparing the placement of atoms, you can determine whether any actual changes were made to the placement of atoms. No atoms should have changed positions. Only the atoms within the missing loops should have been added.
Adding Hydrogens
hbonds in particular and hydrogens in general play an important role in docking calculations, however, due to the resolution limitations of X ray crystallography, hydrogens typically aren't included in a solved protein structure. They will need to be added artificially in Chimera or ChimeraX before docking.
Chimera:
1. Use Tools->Structure Editing->AddH 2. Click OK in the pop-up window
ChimeraX:
1. Type “addh” into the command line.
2. Type "show sidechain" into the command line. The molecule should now look liike the image below, where the white atoms are hydrogens:
This will populate the entire protein with hydrogens, but it will also make the N and C termini charged when we add charges, which isn’t reflective of the charge state of those residues in nature. The receptor in 3WZE is only a portion of the VEGFR II protein, so the backbone ends in 3WZE aren't the protein's true N and C termini. Chimera and ChimeraX has no way of knowing this however, so it will add one too many hydrogens at the N terminal side of the receptor and one too few at the C terminal side of the receptor. This incorrect hydrogenation may not be reflective of the protein in nature, but it is reflective of the protein construct as it was crystallized, so it doesn’t need to be corrected as long as the charged termini aren’t close enough to the active site to mess with docking. In the case of 3WZE, they are not too close, so we can leave the construct's ends as they are.
Adding Charges
Also important for the calculations that go into docking are atomic charges. Files from the PDB won't have assigned charges for each atom, but they'll need to be added for docking.
Chimera:
1. Use Tools->Structure Editing->Add Charge
Standard residues: AMBER ff14SB Other residues: AM1-BCC
2. Click OK in the pop-up window
ChimeraX
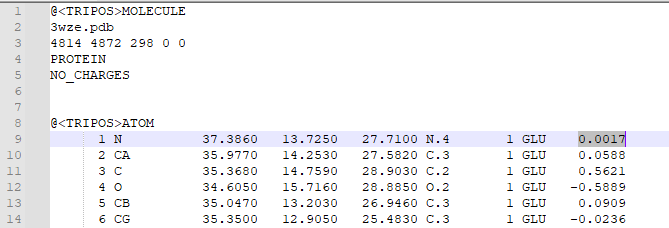
1. Type "addcharge" 2. Save the molecule as a mol2 file 3. Open the mol2 file in a text editor in order to check that charges were added. Atomic charges will appear in the last column of the mol2 file, as highlighted below:
Preparing the Ligand
Chimera:
1. Use Select->Residue->BAX to select the ligand.
2. Use Select->Invert (all models)
3. Use Actions->Atoms/Bonds->Delete
4. Use Tools->Structure Editing->AddH
5. Return to the paper, and check whether the hydrogens added by ChimeraX is accurate to the actual chemistry. Be considerate of any diagrams depicting hbonds or charge-charge interactions when looking for clues. Also be mindful of pH of the solution used to generate crystals. In the case of 3WZE, the hydrogens added by Addh are correct.
6. Save the ligand as a mol2 file.
7. Open it in a text viewer such as Text Editor, or open it from the command line with commands like "vi" or "nano"
8. Check that the bonds are correct. For this particular ligand, bonds 7 and 8 were erroneously labeled as double bonds when they should be amide bonds, and the corresponding Nitrogen atoms (N12 and N14, or atoms 26 and 27 in the mol2 file) are labeled as “N.pl3” instead of “N.am”.
9. Change the “2” in the last column of bond 7’s and bond 8’s rows to “am”. This will change the double bond to an amide bond. Change the “N.pl3” to “N.am” in the rows for atoms 26 and 27. This will make Nitrogen 12 and 14 into amide nitrogens as they should be.
11. Save the new mol2 file
12. Open the mol2 in Chimera
13. Use Tools->Structure Editing->Add Charge to add charges
Standard residues: AMBER ff14SB Other residues: AM1-BCC
14. Click OK in the pop-up window. For 3WZE, an additional pop-up will appear asking you to set the overall charge of the molecule because it isn't a standard molecule that Chimera would know the charge of. Set the charge based on your knowledge of chemistry and the information in the paper. For 3WZE, set it to 0.
15. Save the molecule as a final mol2 file.
ChimeraX:
1. Open the original 3WZE.pdb file in ChimeraX
2. Select the Sorafinib inhibitor by typing "select :BAX". A green outline should appear around the ligand. The non-standard residue name assigned to sorafinib is "BAX" for some reason.
3. Use Select->Invert to select everything except the ligand. Everything except the ligand should now have a green outline.
4. Type "delete sel". You should just be left with your ligand:
5. Type "addh"
6. Return to the paper, and check whether the hydrogens added by ChimeraX is accurate to the actual chemistry. Be considerate of any diagrams depicting hbonds or charge-charge interactions when looking for clues. Also be mindful of pH of the solution used to generate crystals. In the case of 3WZE, the hydrogens added by Addh are correct.
7. Save the ligand as a mol2 file.
8. Open it in a text viewer such as Text Editor, or open it from the command line with commands like "vi" or "nano"
9. Check that the bonds are correct. For this particular ligand, bonds 7 and 8 were erroneously labeled as double bonds when they should be amide bonds, and the corresponding Nitrogen atoms (N12 and N14, or atoms 26 and 27 in the mol2 file) are labeled as “N.pl3” instead of “N.am”.
10. Change the “2” in the last column of bond 7’s and bond 8’s rows (shown below) to “am”. This will change the double bond to an amide bond.
Change the “N.pl3” to “N.am” in the rows for atoms 26 and 27. This will make Nitrogen 12 and 14 into amide nitrogens as they should be.
11. Save the new mol2 file
12. Open the mol2 in ChimeraX
13. With most ligands, you should be able to simply type “addcharge” to assign charges to each atom and save as a mol2 file. However, doing that with this molecule will erroneously assign a +1 charge somewhere and yield an error message. To avoid that, force ChimeraX to assign a neutral charge by typing “addcharge nonstd :BAX BAX 0”
14. Save the molecule as a final mol2 file.
Surface & Spheres
At this stage, move directories to Seawulf.
Surface Generation
The generation of the surface of the receptor protein will have better visualization of the binding site of the receptor. In order to do so, load the mol2 file of 3WZE receptor without hydrogens in Chimera:
Action < Surface < show
Then you will be able to visualize the surface of your protein in Chimera, it will be like something in the image below:
Then we will save the dms(molecular surface) file as 3WZE_rec.dms, which will be used in the next step of spheres generation:
Tools < Structure Editing < Write DMS
Move the generated DMS file to the 002_surface_spheres folder on Seawulf.
Change current working directory in terminal for sphere generation and selection:
cd 002_surface_spheres
ChimeraX:
Unfortunately, ChimeraX cannot save a surface as a .dms file, so this section must be completed in Chimera.
Spheres Generation
First create a new file “INSPH” by typing the following into terminal:
vi INSPH
Note that the name "INSPH" is not arbitrary. Attempting to use a differently named input file will yield an error message.
Type the following information into this file (anything after # are comments that are references for reading only, and should not be putting into the file) :
3WZE_rec.dms #Molecular Surface file R #This line specifies whether to make the spheres in the model’s exterior (R) or interior (L) X #Specifies that the surface points from the dms file should be used for making spheres(?) 0 #Minimum radius between spheres 4.0 #Max radius of each sphere 1.4 #Minimum radius of each sphere 3WZE.sph #Name of the output file that will be made
Then save the file and run the following command on terminal: note that if you want to re-run this command, make sure you remove file “OUTSPH” before your re-run because this file cannot be overwritten. Also remove any temp files that sphgen made. Also bear in mind that sphgen will crash if your protein is too big (greater than ~900 amino acids), so if you're getting an error message, try deleting part of your protein which isn't needed for docking before generating the surface and trying to generate spheres.
sphgen -i INSPH -o OUTSPH
The output file “3WZE.sph” can be visualized by using Chimera, load the output file along with the receptor protein mol2 file, the two structures should be overlapping:
Spheres selection
Since we are only interested in what’s happening in the binding site, we will remove spheres that are far away from the ligand. To do so, use the following command and an output file “selected_spheres.sph” will be generated:
sphere_selector 3WZE.sph 3WZE_lig.mol2 10.0 #sphere_selector [sphere output file] [ligand mol2 file] [radius for distance from ligand]
Open this file in Chimera along with the receptor file, you will see something like this:
Box and Grid
Change the current working directory in terminal for box and grid generation:
cd 003_gridbox
Box generation
First we will generate a box that will be used to generate the grid. To generate a box, we will need to create a new file “showbox.in” by using the following command:
vi showbox.in
Then type the following information into the file (anything after # are comments that are references for reading only, and should not be putting into the file) :
Y #Indicates that we want to make a box 8.0 #Extra length beyond the spheres to be included by the box in all directions ../002_surface_spheres/selected_spheres.sph #Indicates that the box should be constructed to encompass the spheres in this file. 1 #This specifies which cluster of spheres should be used to generate the box. 3WZE.box.pdb #Name of output file
Then, run the following code to get the boxed structure:
showbox < showbox.in
Grid generation
Create a new file “grid.in” by using the following command:
vi grid.in
Then type the following information into the file:
allow_non_integral_charges no compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 allow_non_integral_charges yes bump_filter yes bump_overlap 0.75 receptor_file 3WZE_rec.mol2 box_file 3WZE.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Then generate the grid by typing the following command:
grid -i grid.in -o gridinfo.out
This will generate three files: “gridinfo.out”, “grid.bmp”, and “grid.nrg”. Open “gridinfo.out” and check if the information about the receptor matches the that from the paper and your previous preparations. Also check to make sure there are no error messages at the bottom.
Open "grid.nrg" and receptor file in Chimera, we will be able to see the energy grid of the protein:
Open "grid.bmp" and receptor file in Chimera, we will be able to see the bump grid of the protein:
Energy minimization
Change the current working directory in terminal to do preparation for DOCK:
cd 004_dock
You may want to move over grid.nrg and grid.bmp as well.
To improve the accuracy of DOCK calculation of ligand protein binding, we will have to minimize the potential energy of the ligand. To do so, create a new file “min.in” by using the following command:
vi min.in
Then type the following information into the file:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../000_files/3WZE_lig_wH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../000_files/3WZE_lig_wH.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 3WZE.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
Then run the following command through the use of DOCK:
dock6 -i min.in -o min.out
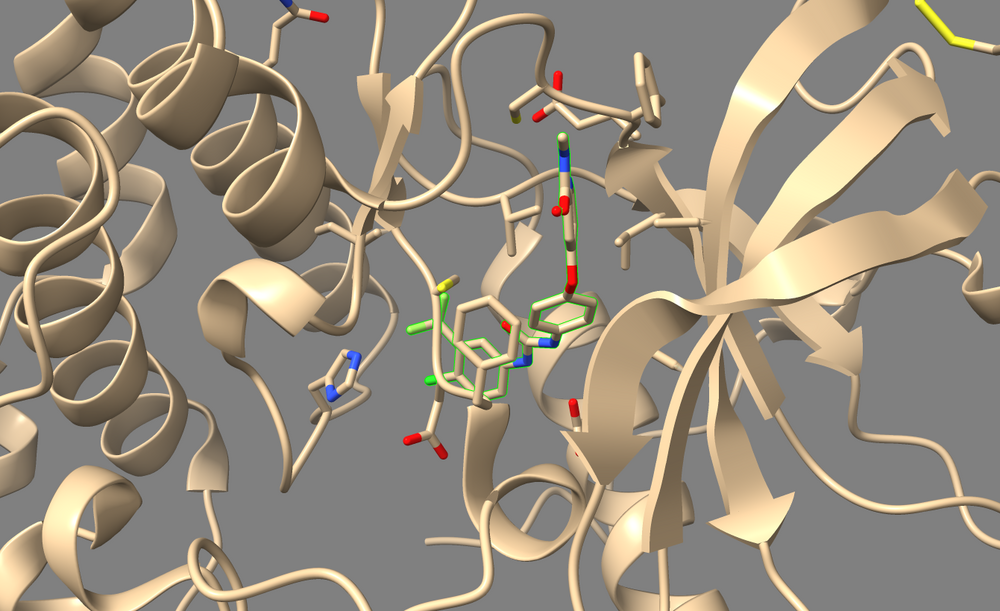
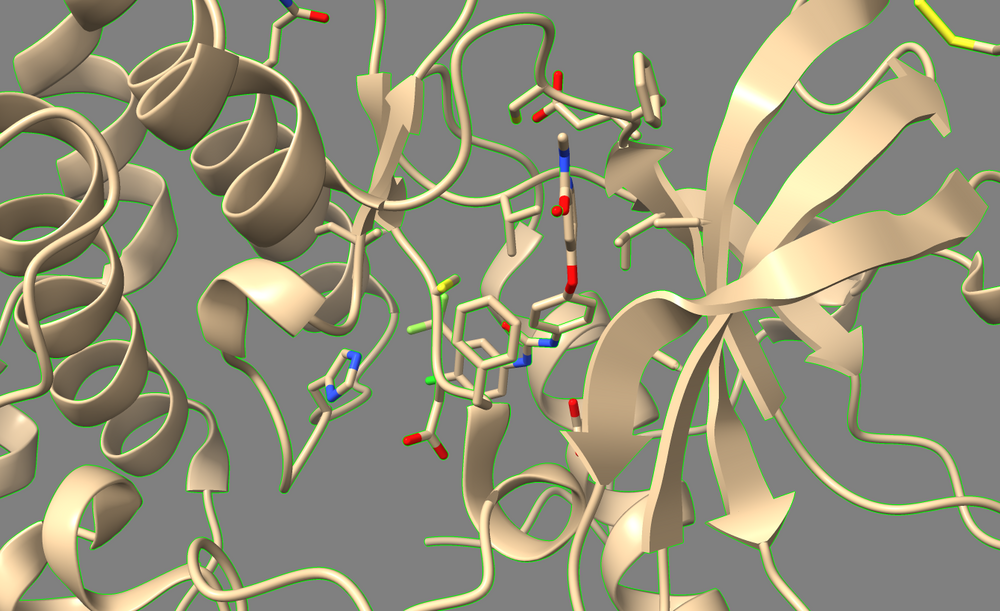
The output file “3WZE.lig.min_scored.mol2”(tan) is the potential energy minimized version of the ligand. Load this file along with ligand mol2 file(blue) in Chimera, you can compare the two molecules:
Reproducing the PDB's binding with DOCK
Change the current directory to perform DOCK:
cd 004_dock
Rigid Docking
This is computationally least expensive since the ligand will be stable, no rotational degrees will be taken into consideration. The energy minimized ligand mol2 file will be used in this simulation. First we will need to create a new file “rigid.in”:
vi rigid.in
Type the following information into the new file:
conformer_search_type rigid use_internal_energy yes ligand_atom_file 3WZE.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 3WZE.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002_surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003_gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 1 rank_ligands no
Then run the following command:
dock6 -i rigid.in -o rigid.out
If the inputs are all correct, there will be a new output file “rigid.out_scored.mol2”.
Fixed Anchor Docking
In fixed anchor docking, most part of the ligand will remain fixed, and the rest will have some rotational freedom. First create a new file “fixed.in”:
vi fixed.in
Then type the following into the file:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001_structure/3WZE_lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001_structure/3WZE_lig.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix fixed.out write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Then run the following command:
dock6 -i fixed.in -o fixed.out
If the inputs are all correct, there will be a new output file “fixed.out_scored.mol2”.
Flexible Docking
Flexible docking is the computationally most expensive out of the three types of docking we introduced. Full rotational freedom and internal degrees of freedom are being considered. First creat a new file “flex.in”:
vi flex.in
Type the following information into the file:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 3WZE.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 3WZE.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002_surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 1 rank_ligands no
Then run the following command:
dock6 -i flex.in -o flex.out
If the inputs are all correct, there will be a new output file “flex.out_scored.mol2”.
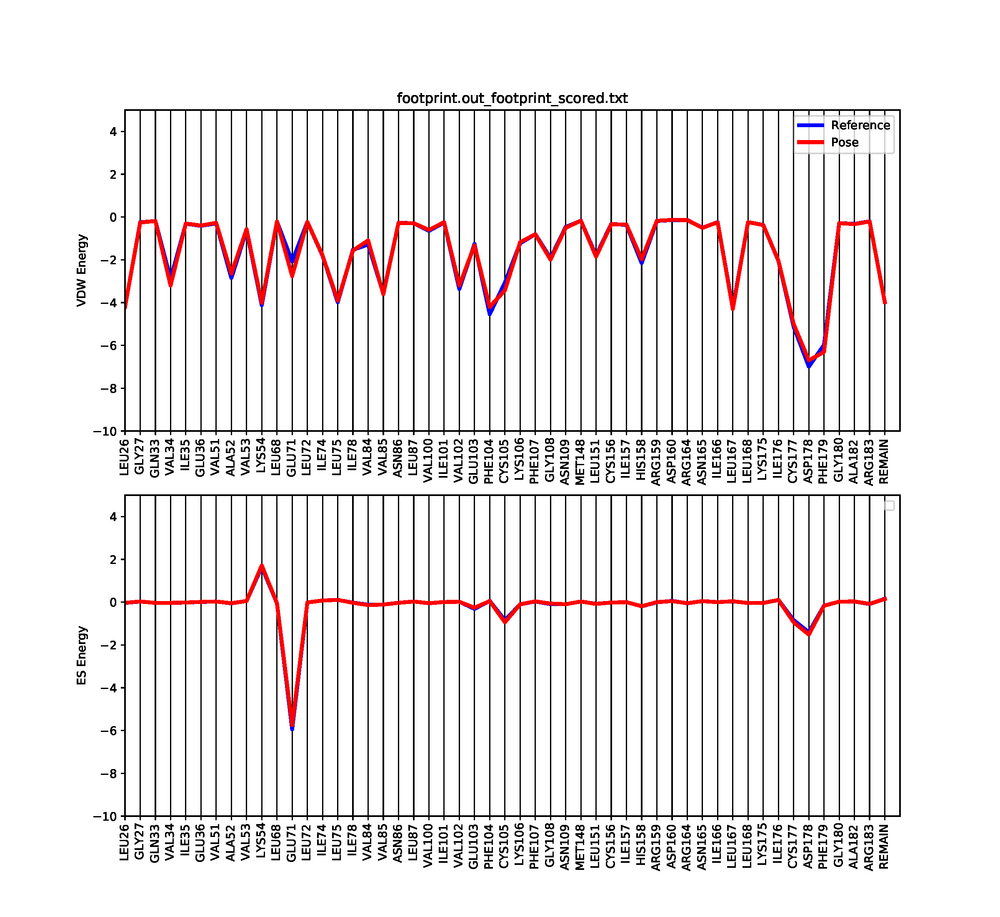
Footprinting
Dock6 has a feature in which we can visualize both the electrostatic and Van der interactions between the ligand and protein. This will show which amino acids are contributing to the binding of the ligand to the protein.
To determine the molecular footprint, create the following input file in your DOCK directory:
cd 004_dock
vi footprint.in
Copy the following into your file:
conformer_search_type rigid use_internal_energy no ligand_atom_file 3WZE.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ../001_structure/3wze.lig.wH.am1bcc.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ../001_structure/3wze_rec_wH.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 9 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix footprint.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
In order to run the footprint calculation, type:
dock6 -i footprint.in -o footprint.out
An error might occur when running footprint with dock, but that is fine. Three files will be generated after a successful run. The files that are generated are footprint.out_footprint_scored.txt footprint.out_hbond_scored.txt footprint.out_scored.mol2. In order to visualize the molecular footprint we need to use a python script provided here a prewritten python script. To run the script just do:
python plot_footprint_single_magnitude.py footprint.out_footprint_scored.txt 50
This python script will generate a graph that shows the Van der Waals and Electrostatic contributions of the 50 most contributing amino acids. Use a pdf viewer to visualize the graph you just created. It should look similar to this:
Virtual Screen
By having a predetermined compound library, virtual screening in DOCK can score these in the compound library based on the input parameters. In this case, we used a smaller library “VS_library_25K.mol2” that was provided by the instructor . Change the current working directory and make sure “VS_library_25K.mol2” is copied to this directory:
cd 005_virtual_screen
First create a new file “virtual.in” for inputting virtual screening parameters:
vi virtual.in
Then type the following information into the file:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file VS_library_25K.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002_surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003_gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix virtual.out write_orientations no num_scored_conformers 1 rank_ligands no
Type the following command:
dock6 -i virtual.in
If the command runs successfully, the file "virtual.in" has set up successfully. We then move to another working directory, and make sure both files "virtual.in" and “VS_library_25K.mol2” is in this directory:
cd 006_virtual_screen_mpi
To prevent overload of the current working load and speed up the process, we can write the code for virtual screening along with requesting more nodes in s shell script and submit the job. To do so, first create a new file “virtual.sh”:
vi virtual.sh
Then type the following information into this file:
#!/bin/bash #SBATCH --time=5:00:00 #SBATCH --nodes=4 #SBATCH --ntasks=112 #SBATCH --job-name=3WZE_flex_dock #SBATCH --output=3WZE_flex_dock #SBATCH -p long-28core mpirun -np 112 dock6.mpi -i virtual.in -o 3WZE.virtual.mpi.out
Then submit the job by typing the following command:
sbatch virtual.sh
Since we specified the number of task to be 112, we are expecting to see 112 output files, name in the format "virtual.out.xxx". In addition, the output contains a mol2 file with all 112 virtual screening results inside.
Cartesian Energy Minimization
Change the current working directory to perform cartesian energy minimization:
cd 007_cartesian_min
We have to perform energy minimization on the virtual screening results before analyzing. First, create a new file “cart_min.in”:
vi cart_min.in
Then type the following information into the file:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../006_virtual_screen_mpi/3WZE_virtual.out_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../001_structure/3WZE_rec.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 3WZE.virtualscreen_cart.min write_orientations no num_scored_conformers 1 rank_ligands no
Then we write a new shell script “cart_min.sh” to request node and run the code to prevent overflow:
vi cart_min.sh
The following lines should be included in the shell file:
#!/bin/bash #SBATCH --time=10:00:00 #SBATCH --nodes=4 #SBATCH --ntasks=112 #SBATCH --job-name=3WZE_flex_dock #SBATCH --output=3WZE_flex_dock #SBATCH -p long-28core mpirun -np 112 dock6.mpi -i cart_min.in -o 3WZE_cart_min.virtual.mpi.out
Then submit the job by running the following command:
sbatch cart_min.sh
We then will be able to see the results.
Rescoring the Virtual Screen
Change the working directory to perform rescoring:
cd 008_rescore
We can rescore the Cartesian energy minimized virtual screening results. This can rank the results based on user defined molecular properties. To do so, make a new file “rescore.in”:
vi rescore.in
Then type the following information into the file:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../007_cartesian_min/3WZE.virtual_screen.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename ../004_dock/3wze.lig.min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../001_structure/3wze_rec.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../004_dock/3wze.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../004_dock/3wze.lig.min_scored.mol2 descriptor_hms_score_ref_filename ../004_dock/3wze.lig.min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename ../004_dock/3wze.lig.min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/ph4.defn ligand_outfile_prefix descriptor.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
To prevent overload, we write a new shell script to request nodes and run the line:
vi rescore.sh
Type the following information into the file:
#!/bin/bash #SBATCH --time=10:00:00 #SBATCH --nodes=4 #SBATCH --ntasks=112 #SBATCH --job-name=3WZE_flex_dock #SBATCH --output=3WZE_flex_dock #SBATCH -p long-28core mpirun -np 112 dock6.mpi -i rescore.in -o rescore.out
Then run the following command:
sbatch rescore.sh
There will be a file “descriptor.out_scored.mol2” when the job is done. We can download the file to local and perform visual analysis by using Chimera.