Difference between revisions of "2024 DOCK GA tutorial 1 with 1NDV"
Stonybrook (talk | contribs) (→I. Introduction) |
Stonybrook (talk | contribs) (→III. Performing GA) |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
A genetic algorithm is a form of de novo design. This form of de novo grows new molecules through a process of evolutionary growth that includes crossover of parent molecules, mutations, and a "natural selection" process. Crossover occurs when two parent molecules have rotatable bonds in the same area of the binding site. This will exchange the fragments between the parents, and creature two new offspring. There are 4 different mutations that can occur during genetic growth. There can be deletion of an existing fragment, addition of a new fragment, substitution of a terminal fragment, and replacement of a linker fragment. After crossover and mutation occurs the new molecules are scored and a select amount are carried over into the next stage of growth. In this tutorial will be using the Elitism elimination method, so only the top scoring molecules will move on to the next generation. | A genetic algorithm is a form of de novo design. This form of de novo grows new molecules through a process of evolutionary growth that includes crossover of parent molecules, mutations, and a "natural selection" process. Crossover occurs when two parent molecules have rotatable bonds in the same area of the binding site. This will exchange the fragments between the parents, and creature two new offspring. There are 4 different mutations that can occur during genetic growth. There can be deletion of an existing fragment, addition of a new fragment, substitution of a terminal fragment, and replacement of a linker fragment. After crossover and mutation occurs the new molecules are scored and a select amount are carried over into the next stage of growth. In this tutorial will be using the Elitism elimination method, so only the top scoring molecules will move on to the next generation. | ||
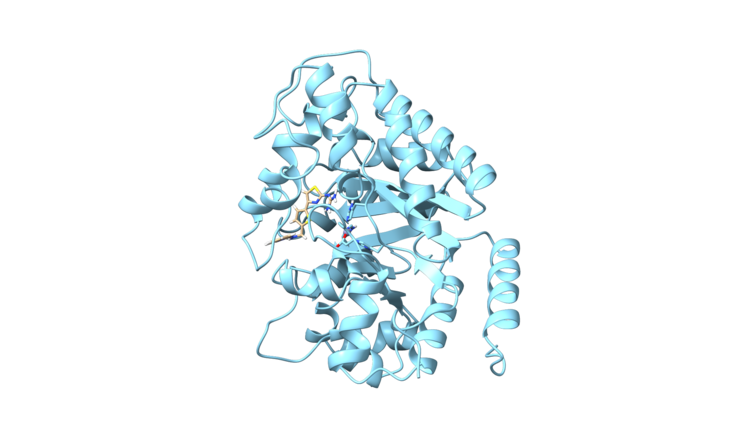
| − | [[File:1ndv_original.png|center|thumb| | + | [[File:1ndv_original.png|center|thumb|750px|PDB 1NDV]] |
=='''II. Fragment Library Generation for 1NDV'''== | =='''II. Fragment Library Generation for 1NDV'''== | ||
| − | + | In order for the GA to work, we first need to produce a large and diverse fragment library. Make a directory to run the GA in called "1NDV_GA". Now enter that directory and make a new directory named "fraglib". This is where we will build our fragment library. Create a new file for the fragment library and use it as input for dock6 so we can interactively build the library using the following commands. | |
| + | |||
| + | touch 1NDV.fraglib | ||
| + | dock6 -i 1NDV.fraglib | ||
| + | |||
| + | Now answer the prompts using the responses below as a guide. | ||
| + | |||
| + | conformer_search_type flex | ||
| + | write_fragment_libraries yes | ||
| + | fragment_library_prefix 1ndv.fraglib | ||
| + | fragment_library_freq_cutoff 1 | ||
| + | fragment_library_sort_method freq | ||
| + | fragment_library_trans_origin no | ||
| + | use_internal_energy no | ||
| + | ligand_atom_file /gpfs/projects/AMS536/2024/students/group_2_1NDV/dock_screen/001.structure/1ndv_ligand_only_H_charge.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 1ndv.frag.output | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Now combine the torsion environment file (1NDV_torenv.dat) with the full_sorted_fraglib.dat. This can be done with the python script combine_torenv.py using the following command. The new torenv.dat will be used for the GA. | ||
| + | |||
| + | python ${DOCK_DIR}/bin/combine_torenv.py 2NNQ.fraglib_torenv.dat ${DOCK_DIR}/parameters/fraglib_torenv.dat | ||
=='''III. Performing GA'''== | =='''III. Performing GA'''== | ||
| + | It is now time to perform the GA. Let's make a new directory to store our results called "1NDV_GA_results". Enter this new directory and create a new file for the input of our GA called 1NDV_GA.in. It is recommended that you input the parameters interactively as we did before. To do this, simply give dock6 the empty input file and follow the inputs below as a guide. Things such as the number of generations, elimination method, and mutations allowed can be adjusted to suit your needs. Do not submit this job! We will create a slurm job in the next step. | ||
| + | conformer_search_type genetic | ||
| + | ga_molecule_file /Path/1ndv_ligand_only_H_charge.mol2 | ||
| + | ga_utilities no | ||
| + | ga_fraglib_scaffold_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_ga_scaffold.mol2 | ||
| + | ga_fraglib_linker_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 | ||
| + | ga_fraglib_sidechain_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 | ||
| + | ga_torenv_table ../fraglib/unique_full_sorted_fraglib.dat | ||
| + | ga_max_generations 100 | ||
| + | ga_xover_on yes | ||
| + | ga_xover_sampling_method_rand yes | ||
| + | ga_xover_max 150 | ||
| + | ga_bond_tolerance 0.5 | ||
| + | ga_angle_cutoff 0.14 | ||
| + | ga_check_overlap no | ||
| + | ga_mutate_addition yes | ||
| + | ga_mutate_deletion yes | ||
| + | ga_mutate_substitution yes | ||
| + | ga_mutate_replacement yes | ||
| + | ga_mutate_parents yes | ||
| + | ga_pmut_rate 0.3 | ||
| + | ga_omut_rate 0.7 | ||
| + | ga_max_mut_cycles 5 | ||
| + | ga_mut_sampling_method rand | ||
| + | ga_num_random_picks 10 | ||
| + | ga_max_root_size 5 | ||
| + | ga_energy_cutoff 100 | ||
| + | ga_heur_unmatched_num 2 | ||
| + | ga_heur_matched_rmsd 2 | ||
| + | ga_constraint_mol_wt 550 | ||
| + | ga_constraint_rot_bon 10 | ||
| + | ga_constraint_H_accept 10 | ||
| + | ga_constraint_H_don 5 | ||
| + | ga_constraint_formal_charge 4 | ||
| + | ga_ensemble_size 200 | ||
| + | ga_selection_method elitism | ||
| + | ga_elitism_combined no | ||
| + | ga_elitism_option max | ||
| + | ga_max_num_gen_with_no_crossover 1000 | ||
| + | ga_name_identifier ga | ||
| + | ga_output_prefix 1NDV_GA_output | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100 | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary no | ||
| + | multigrid_score_primary no | ||
| + | dock3.5_score_primary no | ||
| + | continuous_score_primary no | ||
| + | footprint_similarity_score_primary no | ||
| + | pharmacophore_score_primary no | ||
| + | hbond_score_primary no | ||
| + | internal_energy_score_primary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_use_grid_score yes | ||
| + | descriptor_use_pharmacophore_score no | ||
| + | descriptor_use_tanimoto no | ||
| + | descriptor_use_hungarian no | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_grid_score_rep_rad_scale 1 | ||
| + | descriptor_grid_score_vdw_scale 1 | ||
| + | descriptor_grid_score_es_scale 1 | ||
| + | descriptor_grid_score_grid_prefix grid | ||
| + | descriptor_volume_score_reference_mol2_filename descriptor_volume_score_reference.mol2 | ||
| + | descriptor_volume_score_overlap_compute_method analytical | ||
| + | descriptor_weight_grid_score 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1 | ||
| + | simplex_trans_step 1 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 00 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min yes | ||
| + | simplex_coefficient_restraint 10 | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/chem.defn | ||
| + | |||
| + | To create a slurm submission script, first create a file called "1NDV_GA.sh". Input the following lines into the new file. | ||
| + | |||
| + | #!/bin/bash | ||
| + | # | ||
| + | #SBATCH --job-name=1ndv_GA | ||
| + | #SBATCH --output=GA.txt | ||
| + | #SBATCH --ntasks-per-node=24 | ||
| + | #SBATCH --nodes=6 | ||
| + | #SBATCH --time=1:00:00 | ||
| + | #SBATCH -p debug-28core | ||
| + | |||
| + | module load intel/mpi/64/2018/18.0.3 | ||
| + | |||
| + | mpirun -np 144 dock6.mpi -i 1NDV_GA.in -o 1NDV_GA.out | ||
| + | |||
| + | You can now sumbit the job using: | ||
| + | sbatch 1NDV_GA.sh | ||
| + | |||
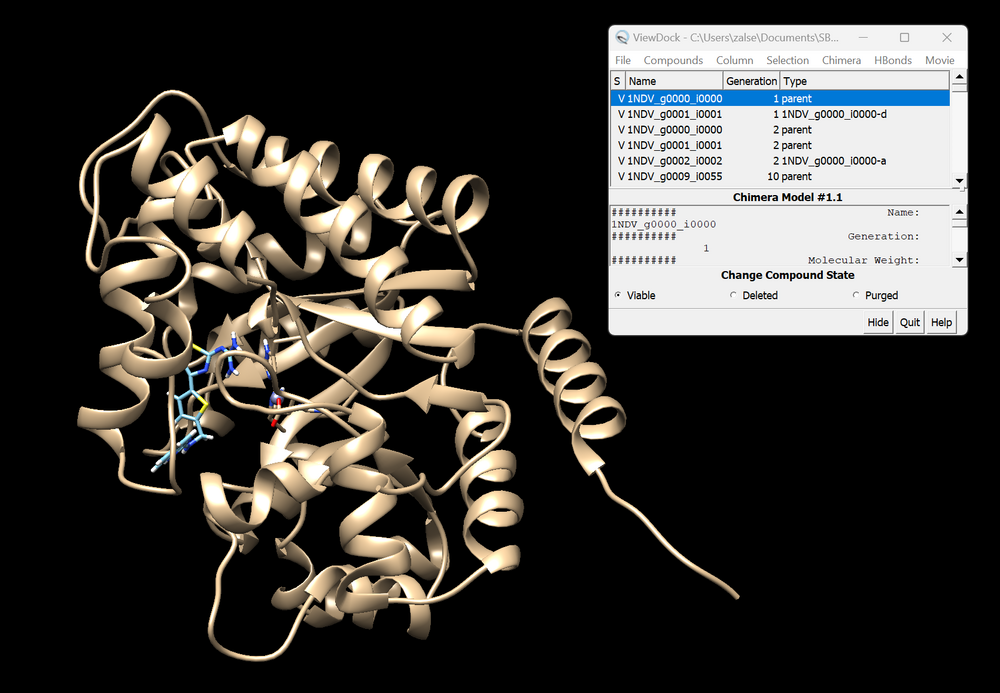
| + | The GA will take some time to run, but be sure to check that you do not have any errors early on. Once the GA has run, you should now have a directory with mol2 files for every generation of the GA. You can download all of these individually, but it may be easier to use the "cat" command to place them into one file. Once you have your desired generations downloaded on your local computer, start a new Chimera session. (Note that his works best on Chimera and not Chimera X) Start by opening the mol2 file that only contains the protein. Now navigate to the tool bar click "Tools" → "Surface/Binding Analysis" → "View Dock". Open the mol2 files each generation of GA. Be sure to select DOCK6 as your file type. You can then use the ViewDock toolbar to select "Column" → "Show" → "Generation" and "Column" → "Show" → "Type". This will show what generation each ligand is from, and what the crossover or mutation was that generated the structure. An example of this is shown below. | ||
| + | [[File:1ndv_ga.png|center|1000px|PDB 1NDV]] | ||
Latest revision as of 07:46, 6 May 2024
I. Introduction
A genetic algorithm is a form of de novo design. This form of de novo grows new molecules through a process of evolutionary growth that includes crossover of parent molecules, mutations, and a "natural selection" process. Crossover occurs when two parent molecules have rotatable bonds in the same area of the binding site. This will exchange the fragments between the parents, and creature two new offspring. There are 4 different mutations that can occur during genetic growth. There can be deletion of an existing fragment, addition of a new fragment, substitution of a terminal fragment, and replacement of a linker fragment. After crossover and mutation occurs the new molecules are scored and a select amount are carried over into the next stage of growth. In this tutorial will be using the Elitism elimination method, so only the top scoring molecules will move on to the next generation.
II. Fragment Library Generation for 1NDV
In order for the GA to work, we first need to produce a large and diverse fragment library. Make a directory to run the GA in called "1NDV_GA". Now enter that directory and make a new directory named "fraglib". This is where we will build our fragment library. Create a new file for the fragment library and use it as input for dock6 so we can interactively build the library using the following commands.
touch 1NDV.fraglib dock6 -i 1NDV.fraglib
Now answer the prompts using the responses below as a guide.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix 1ndv.fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy no ligand_atom_file /gpfs/projects/AMS536/2024/students/group_2_1NDV/dock_screen/001.structure/1ndv_ligand_only_H_charge.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1ndv.frag.output write_orientations no num_scored_conformers 1 rank_ligands no
Now combine the torsion environment file (1NDV_torenv.dat) with the full_sorted_fraglib.dat. This can be done with the python script combine_torenv.py using the following command. The new torenv.dat will be used for the GA.
python ${DOCK_DIR}/bin/combine_torenv.py 2NNQ.fraglib_torenv.dat ${DOCK_DIR}/parameters/fraglib_torenv.dat
III. Performing GA
It is now time to perform the GA. Let's make a new directory to store our results called "1NDV_GA_results". Enter this new directory and create a new file for the input of our GA called 1NDV_GA.in. It is recommended that you input the parameters interactively as we did before. To do this, simply give dock6 the empty input file and follow the inputs below as a guide. Things such as the number of generations, elimination method, and mutations allowed can be adjusted to suit your needs. Do not submit this job! We will create a slurm job in the next step.
conformer_search_type genetic ga_molecule_file /Path/1ndv_ligand_only_H_charge.mol2 ga_utilities no ga_fraglib_scaffold_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_ga_scaffold.mol2 ga_fraglib_linker_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 ga_fraglib_sidechain_file /gpfs/projects/rizzo/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 ga_torenv_table ../fraglib/unique_full_sorted_fraglib.dat ga_max_generations 100 ga_xover_on yes ga_xover_sampling_method_rand yes ga_xover_max 150 ga_bond_tolerance 0.5 ga_angle_cutoff 0.14 ga_check_overlap no ga_mutate_addition yes ga_mutate_deletion yes ga_mutate_substitution yes ga_mutate_replacement yes ga_mutate_parents yes ga_pmut_rate 0.3 ga_omut_rate 0.7 ga_max_mut_cycles 5 ga_mut_sampling_method rand ga_num_random_picks 10 ga_max_root_size 5 ga_energy_cutoff 100 ga_heur_unmatched_num 2 ga_heur_matched_rmsd 2 ga_constraint_mol_wt 550 ga_constraint_rot_bon 10 ga_constraint_H_accept 10 ga_constraint_H_don 5 ga_constraint_formal_charge 4 ga_ensemble_size 200 ga_selection_method elitism ga_elitism_combined no ga_elitism_option max ga_max_num_gen_with_no_crossover 1000 ga_name_identifier ga ga_output_prefix 1NDV_GA_output use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary no multigrid_score_primary no dock3.5_score_primary no continuous_score_primary no footprint_similarity_score_primary no pharmacophore_score_primary no hbond_score_primary no internal_energy_score_primary no descriptor_score_primary yes descriptor_use_grid_score yes descriptor_use_pharmacophore_score no descriptor_use_tanimoto no descriptor_use_hungarian no descriptor_use_volume_overlap yes descriptor_grid_score_rep_rad_scale 1 descriptor_grid_score_vdw_scale 1 descriptor_grid_score_es_scale 1 descriptor_grid_score_grid_prefix grid descriptor_volume_score_reference_mol2_filename descriptor_volume_score_reference.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_grid_score 1 descriptor_weight_volume_overlap_score -1 minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 00 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/chem.defn
To create a slurm submission script, first create a file called "1NDV_GA.sh". Input the following lines into the new file.
#!/bin/bash # #SBATCH --job-name=1ndv_GA #SBATCH --output=GA.txt #SBATCH --ntasks-per-node=24 #SBATCH --nodes=6 #SBATCH --time=1:00:00 #SBATCH -p debug-28core
module load intel/mpi/64/2018/18.0.3 mpirun -np 144 dock6.mpi -i 1NDV_GA.in -o 1NDV_GA.out
You can now sumbit the job using:
sbatch 1NDV_GA.sh
The GA will take some time to run, but be sure to check that you do not have any errors early on. Once the GA has run, you should now have a directory with mol2 files for every generation of the GA. You can download all of these individually, but it may be easier to use the "cat" command to place them into one file. Once you have your desired generations downloaded on your local computer, start a new Chimera session. (Note that his works best on Chimera and not Chimera X) Start by opening the mol2 file that only contains the protein. Now navigate to the tool bar click "Tools" → "Surface/Binding Analysis" → "View Dock". Open the mol2 files each generation of GA. Be sure to select DOCK6 as your file type. You can then use the ViewDock toolbar to select "Column" → "Show" → "Generation" and "Column" → "Show" → "Type". This will show what generation each ligand is from, and what the crossover or mutation was that generated the structure. An example of this is shown below.