Difference between revisions of "2024 DOCK tutorial 2 with PDBID 1NDV"
Stonybrook (talk | contribs) (→Protein Prep) |
Stonybrook (talk | contribs) (→Protein Prep) |
||
| Line 71: | Line 71: | ||
6. We also need to generate a .mol2 file. So go to File → Save Mol2 and give it a name 1ndv_protein_only.mol2. The previous file as a .pdb extension and will not be overwritten as a mol2 file is separate from mol2 format. | 6. We also need to generate a .mol2 file. So go to File → Save Mol2 and give it a name 1ndv_protein_only.mol2. The previous file as a .pdb extension and will not be overwritten as a mol2 file is separate from mol2 format. | ||
| + | |||
| + | ====Adding Hydrogens and Charges==== | ||
We need to prepare the protein with hydrogens and charges. We must add hydrogens before adding charges. | We need to prepare the protein with hydrogens and charges. We must add hydrogens before adding charges. | ||
Revision as of 11:47, 6 May 2024
Contents
Introduction
DOCK is a powerful computational tool used in drug design. "Docking" refers to the identification of low energy binding conformations of a small molecule, or ligand, in an active site. There are three methods in which DOCK performs docking: rigid, fixed anchor, and flexible. DOCK also has the ability to perform a virtual screen, which is analogous to a high throughput screen often performed in wet labs. This method allows for thousands of molecules to be docked and ranked so that the highest scoring molecules can move to the next stage of testing. A virtual screen can save both time and money when compared to a traditional high throughput screen. This tutorial will guide you through the process of performing each method of docking, a virtual screen, and associated analysis using PDB 1NDV on a HPC cluster (Seawulf). Replace 1NDV with your target protein in this tutorial.
Required Software and Skills
In order to complete this tutorial UCSF Chimera is required. Chimera is a powerful visualization tool that will aid in the preparation and analysis of the virtual screen. Please note that the newer version, Chimera X, can not be used for this tutorial as it lacks certain integrations with DOCK. While this tutorial will walk you through all steps necessary to use Chimera, it may be helpful to view this tutorial for a more in depth guide on the program.
It is also recommended that you have experience with Unix and vi. A Unix tutorial can be found on the site here and a vi tutorial can be found here.
Set-up
Before we begin, we need to set up our directories and download our file from the Protein Data Bank (PDB).
Creating Directories
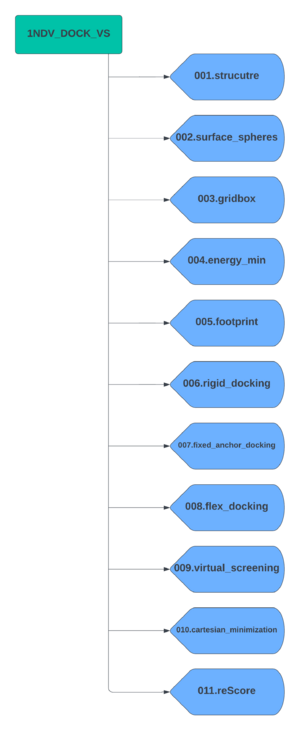
Create a parent directory which will contain all subdirectors for each docking step. For this tutorial we will name the parent directory 1NDV_DOCK_VS. Now create the subdirectories using the following scheme.
Downloading PDB File
Visit the RCSB Protein Data Bank (PDB) website and use the search bar to search for your target protein. As stated before, we will be using 1NDV for this tutorial.
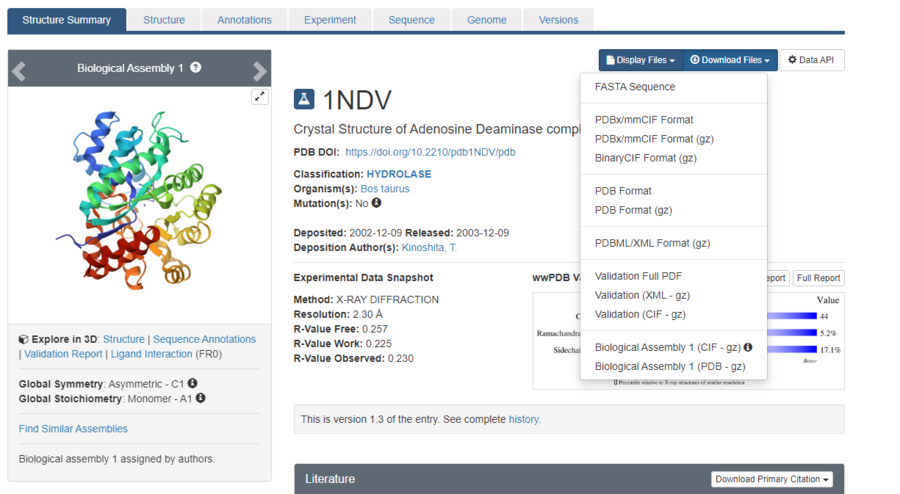
The page for the protein will look like this. Click the "Download Files" dropdown menu, and download the "PDB Format".
With the PDB file saved, we are now ready to begin generating files for DOCK!
Preparing Structure Files
To prepare structure files, the protein and ligand structures need to be prepped. It is important prepare your structure files using the following steps to ensure your files run properly and are considered correct chemical models of your system.
We will prep the protein and ligand structures using Chimera.
Protein
Required file: *Structure file from the RCSB Protein Data Bank (PDB). *For this tutorial, is it 1ndv.pdb.
Analyzing the Protein Structure:
Open 1ndv.pdb in Chimera.
Assess the protein structure:
1. Are there any missing loops?
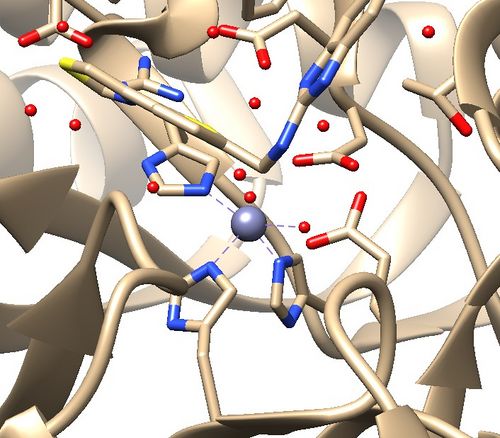
2. Is there a metal coordination atom forming key interactions with the protein?
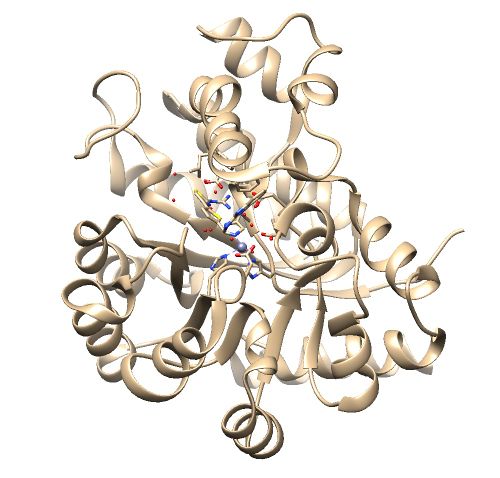
The .pdb for 1ndv from the RCSB Protein Data Bank is shown below.
Assessment:
1. No missing loops.
2. Zinc coordination with the protein and a water molecule. This zinc atom must be kept in the protein prep.
Protein Prep
1. Select an atom on the protein
2. Select the entire protein by pressing the "up" arrow key. You will want to keep the Zn in the complex so hold down the "shift" key and click the Zn atom to select it as well.
3. Go to the menu bar Select → Invert(all models). This will invert the selection and select waters, the ligand, and other compounds in the crystal structure that wasn't previously selected.
4. Go to the menu bar Actions → Atoms/Bonds → Delete.
5. Then save this structure with a a file name (i.e. 1ndv_protein_only.pdb). Go to the menu bar File → Save PBD. Your pdb file will now look similar to this:
6. We also need to generate a .mol2 file. So go to File → Save Mol2 and give it a name 1ndv_protein_only.mol2. The previous file as a .pdb extension and will not be overwritten as a mol2 file is separate from mol2 format.
Adding Hydrogens and Charges
We need to prepare the protein with hydrogens and charges. We must add hydrogens before adding charges.
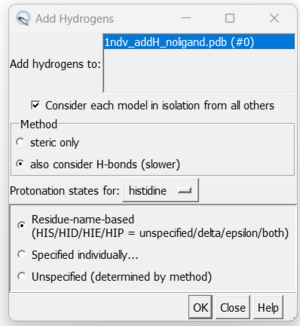
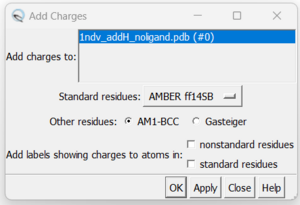
7. Adding hydrogens. To add hydrogen atoms click on: Tools → Structure Editing → AddH. This command will cause the following dialogue box to appear:
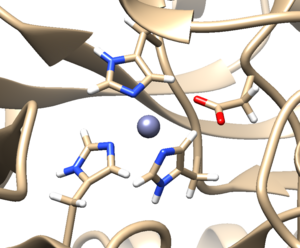
a. Click 'OK'. When the program is finished, the bottom left-hand side will say "Hydrogens added". You may not see any changes in the structure. Here we can see hydrogens on residues surround the zinc atom. If so you can make sure by selecting a couple residues and clicking Actions → Atoms/Bonds → Show. You can unselect these residues by clicking on the background.
b. Now once this check is done save your protein file with hydrogens as a .pdb (i.e 1ndv_protein_H.pdb) and .mol2 (i.e 1ndv_protein_H.mol2). It is useful to save changes in Chimera in steps as each change is not able to be undone.
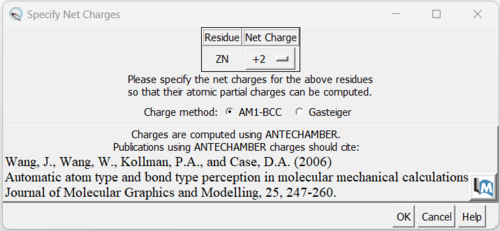
8. Now we can add charges by Tools → Structure Editing → Add Charge and the following dialogue box will show up:
You can keep the defaults selected and click "OK". If you have a metal coordination atom in your structure Chimera will ask you to specify the atom's charge.
Once the program is finished in the bottom left hand corner will display what the total charge of the structure is. This number should be an integer.
The protein structure is now completely prepared and ready for docking.
The final step is to save two files, a .pdb and a .mol2 file of this structure (i.e. 1ndv_protein_H_charges.pdb and 1ndv_protein_H_charges.mol2)
Ligand
We must first create a .pdb file with only the ligand. To do this we must select the ligand in Chimera. There are two ways to do this:
1.Select an atom on the ligand, and use the up arrow until the entire ligand is selected
-or-
2. Use the toolbar and click "Select" → "Structure" → "Ligand"
Once the ligand is selected use "Select" → "Invert" to select all other atoms present in the session.
Then use "Actions" → "Atoms/Bonds" → "Delete" to delete everything except the ligand.
It should look like this.
You can now save only the ligand in a separate .pbd file. We will name this file 1ndv_ligand_only.pdb.
We will also save the ligand in this state as a .mol2 file. We will name this file 1ndv_no_hydrogens_no_charges.mol2
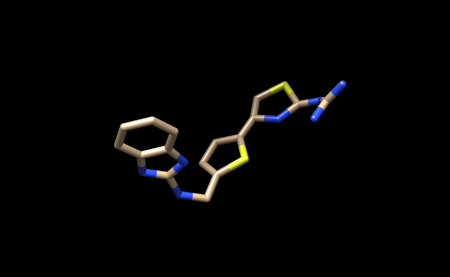
We now need to add hydrogens and charges like we did previously for the protein. It is important to be careful and to cross reference the structure of the ligand in the PDB to ensure that hydrogens are being added in the right places, and that the overall charge of the ligand remains the same. After adding hydrogens, your structure should look similar to this:
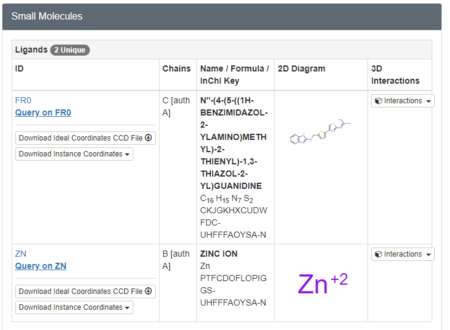
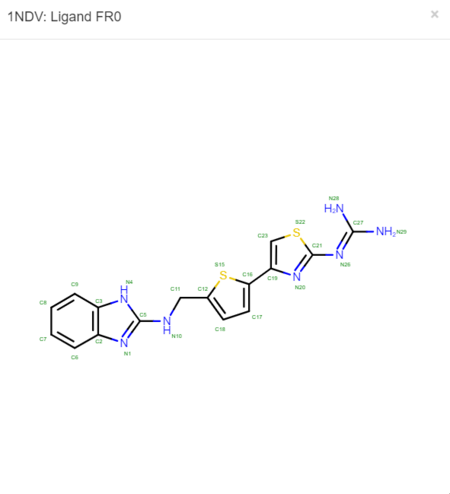
We must now look at the structure in the PDB to ensure Chimera is adding the hydrogens in the proper place. To do this, scroll down on the PDB page of the protein until you see the table labeled "Small Molecules".
Now click on the "2D Diagram" of the ligand to bring up a larger picture.
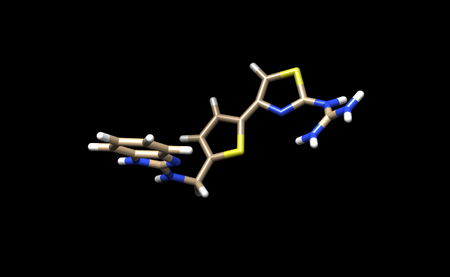
It appears that Chimera has added an extra Hydrogen to N26. We need to remove that by simply selecting and deleting that hydrogen. The ligand should now look like this:
We can now add charges to the ligand. When the dialogue box appears, be sure to set the overall charge to 0 for this ligand. With this step complete we can now save a .pdb file and .mol2 with the name 1ndv_ligand_H_charge.
Upload all four of the .mol2 files created during the protein and ligand preparation process to Seawulf for use with DOCK.
Surface Spheres
DMS file
sphgen
sphere selector
Gridbox
While it is possible to calculate interactions between the ligand and the entire protein surface, this is computationally expensive. In this tutorial we will define a box of interest in the active site of the protein, and DOCK will generate a grid to calculate interaction energies within this box.
Generating the Box
To generate the box we use a DOCK program called showbox. This program is accessed through the command line. First navigate to your 003.gridbox directory and create a new file called showbox.in. Place the following commands into the new file:
Y 8.0 ../002.surface_spheres/selected_spheres.sph 1 1ndv.box.pdb
Remember to change "1ndv" to your protein name. This file may need to be modified for your system if you need a larger box. This file currently draws a box of 8.0 angstroms around the selected spheres. To change this, simply change the "8.0" in the input file.
To run this file type:
showbox < showbox.in
If showbox ran successfully you will now have a file named 1ndv.box.pdb in the directory.
Generating the Grid
Now that the box has been defined, we can generate the grid. We do this using a Dock program called grid. Make a new file called grid.in. Place the following commands into the new file:
allow_non_integral_charges no compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4. bump_filter yes bump_overlap 0.75 receptor_file ../001.structure/1ndv_protein_only_H_charge.mol2 box_file 1ndv.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Be sure you are using the receptor_file and box_file for your protein.
Run the file with the following command:
grid -i grid.in -o 1ndvGridInfo.out
This program will take a few minutes to run. If it ran succesfully you will see three new files in your direcotry:
- grid.bmp
- grid.nrg
- 1ndvGridInfo.out
After successfully generating the box and grid, we can begin energy minimization.
Ligand Energy Minimization
DOCK calculates interaction energies between the protein and the ligand. It is important that we first minimize the ligand to a low energy conformer before we dock it into the binding site of the protein to ensure accurate results.
Frist navigate to the 004.minization directory and create a new file called min.in. Place the follow lines in the new file.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file /gpfs/projects/AMS536/2024/students/group_2_1NDV/dock_screen/001.structure/1ndv_ligand_only_H_charge.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename /gpfs/projects/AMS536/2024/students/group_2_1NDV/dock_screen/001.structure/1ndv_ligand_only_H_charge.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix /gpfs/projects/AMS536/2024/students/group_2_1NDV/dock_screen/003.gridbox/grid minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1ndv.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
Now run the file with the following command:
dock6 -i min.in -o min.out
If the program ran successfully you will see a 1ndv.lig.min.mol2 file.
Save this file to your local computer and open it in Chimera with the original ligand file. You will see that DOCK has made very slight changes to the strucutre.
Footprint Generation
Rigid Docking
The first docking method we will be using is rigid docking. In this method, the ligand is treated as a rigid structure. DOCK attempts to fit the ligand into the binding site using the structure provided, and different ligand conformations are not sampled. For this step we will be working in the 0006.rigid_docking directory and will be using the minimized ligand structure.
Create a new file called rigid.in and place the following lines into it:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/1ndv.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.energy_min/1ndv.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 10 write_conformations yes cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Remember to change paths so that they point to the files on your system. Now run the program using the following command:
dock6 -i rigid.in -o rigid.out
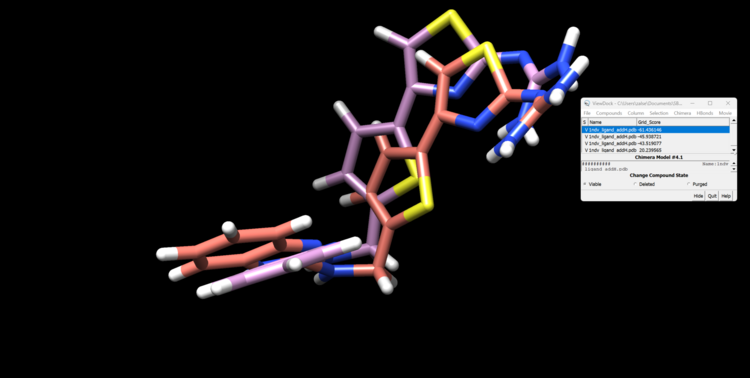
This step may take a few minutes. You will know it ran successfully when you see a file named rigid.out_scored.mol2. This file will contain your docked ligand in 10 orientations. Let's open this file with the minimized ligand file.
First, open the minimized ligand file in Chimera normally. We will use the View Dock tool in Chimera to view the different orientations of the rigid docking results. To do this click "Tools"→ "Surface/Binding Analysis" → "ViewDock". then select "Dock 4,5 or 6" as the file type. Now select "Column" → "Show" → "Grid Score" so we can see how DOCK scored these orientations. You can now view each orientation individually or a selection of them all at once. Here is our top scoring orientation and our minimized ligand.
Fixed Anchor Docking
Flex Docking
The last docking method will we try is called flexible docking. This method is the most computationally expensive, but also samples the most conformational space. In this method DOCK will sample conformations of all rotatable bonds and will attempt to minimize the energy of the ligand. Navigate to the 008.flex_docking directory and create a new file with the name flex.in. Add the following lines to the new file:
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/1ndv.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename
../004.energy_min/1ndv.lig.min_scored.mol2
use_database_filter no orient_ligand yes automated_matching yes receptor_site_file
../002.surface_spheres/selected_spheres.sph
max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no minimize_flexible_growth_ramp no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 1 rank_ligands no
This file will only save 1 conformation. This can be changed by adjusting the "write_orientations" parameter. Use the following command to run the docking:
dock6 -i flex.in -o flex.out
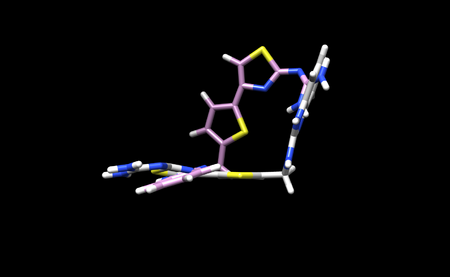
You will know the program ran successfully when you se a flex.out_scored.mol2 file. Save this file on your local system and open it in Chimera along with the minimized ligand file. We can open both of these files normally as we only saved 1 conformation. This is what our docking result looks like in Chimera:
Virtual Screening
Slurm
Cartesian Minimization
In this step we will minimize all the molecules that were docked during the virtual screen. Navigate to the 010.cartesian_minimization directory and create a new file called min.in. Add the following lines to the new file:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../009.virtual_screening/virtual.out.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary no multigrid_score_primary no dock3.5_score_primary no continuous_score_primary yes cont_score_rec_filename ../001.structure/1ndv_protein_only_H_charge.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1ndv.virtual_screen.min write_orientations no num_scored_conformers 1 rank_ligands no
Do NOT run this on the login node. Create a slurm script similar to this one which we tiltled run.min:
#!/bin/bash # #SBATCH --job-name=1ndv_minimization_tutorial #SBATCH --output=vs_output.txt #SBATCH --ntasks=28 #SBATCH --nodes=6 #SBATCH --time=48:00:00 #SBATCH -p long-28core
dock6 -i min.in -o min.out
Once this is complete, we will rescore the molecules based on their minimized structures.