Difference between revisions of "2025 DOCK tutorial 1 with PDBID 1O86"
Stonybrook (talk | contribs) (→001: Structure Prep) |
Stonybrook (talk | contribs) (→002: Spheres) |
||
| Line 36: | Line 36: | ||
== 002: Spheres == | == 002: Spheres == | ||
surface generation, sphgen, selecting spheres, visualization in Chimera | surface generation, sphgen, selecting spheres, visualization in Chimera | ||
| + | '''Generate the required surface file''' | ||
| + | |||
| + | 1. Open 1O86 protein only file in chimera and hit select > show> surface | ||
| + | 2. Write the DMS file by choosing tools>structure editing>Write DMS | ||
| + | 3. Upload the DMS file to your directory | ||
| + | 4. Create a sphere input file using the following command: | ||
| + | vi INSPH | ||
| + | |||
| + | 5. Paste the following into your file: | ||
| + | ./IO86.dms | ||
| + | R | ||
| + | X | ||
| + | 0.0 | ||
| + | 4.0 | ||
| + | 1.4 | ||
| + | IO86.sph | ||
| + | |||
| + | 6. Run the program with the following command | ||
| + | sphgen -i INSPH -o OUTSPH | ||
== 003: Grid/box == | == 003: Grid/box == | ||
Revision as of 14:01, 19 February 2025
Contents
DOCK Tutorial using PDB 1O86
[intro text]
000: Foundations
Directory setup, copying files to Seawulf, Chimera, basic Unix commands
001: Structure Prep
Download PDB, separate lig/rec, model loops, addH/charge
Downloading the PDB
Having setup your necessary environment to work on seawulf, lets navigate to your local computer and begin the protein preparation process:
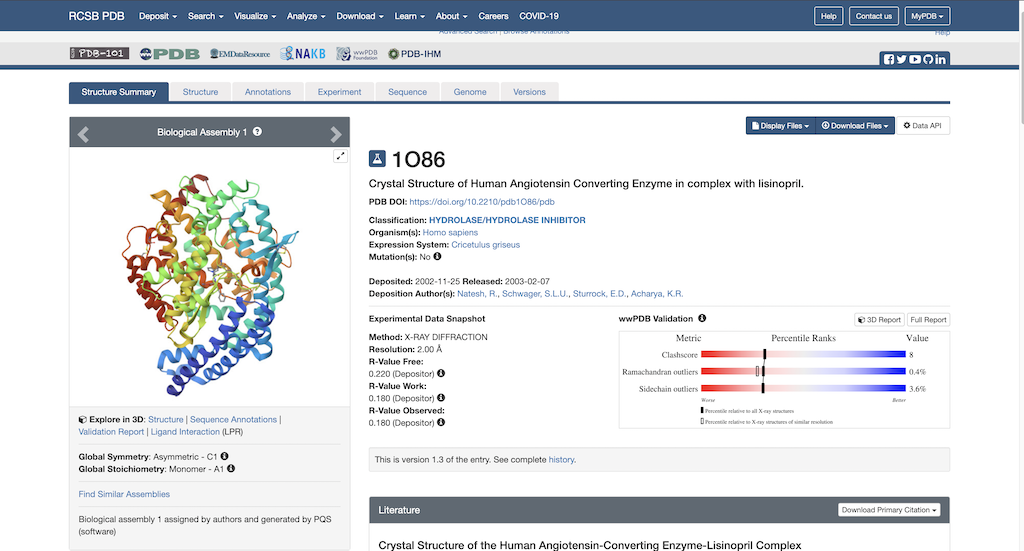
To begin protein preparation you will need the necessary PDB file to work with. Using this link: https://www.rcsb.org/structure/1O86, you will see the RCSB main page opened to our protein of choice:
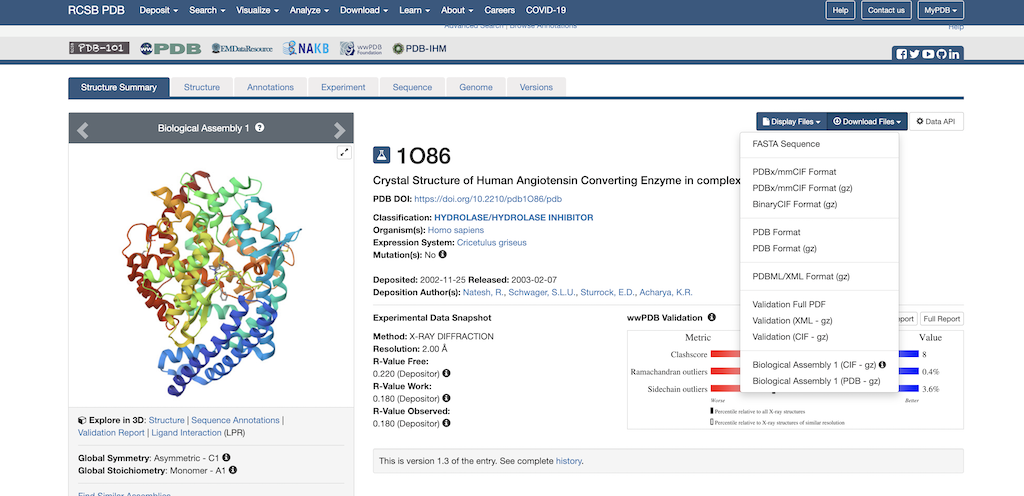
Next, you'll want to navigate to the top right corner where it says Download Files. Then, select the dropdown arrow. The following pulldown menu will appear on the screen:
Select 'Download PDB'. Now the PDB file is downloaded to your local computer.
Now that you have the PDB file, lets navigate to Chimera program to open the file.
002: Spheres
surface generation, sphgen, selecting spheres, visualization in Chimera Generate the required surface file
1. Open 1O86 protein only file in chimera and hit select > show> surface 2. Write the DMS file by choosing tools>structure editing>Write DMS 3. Upload the DMS file to your directory 4. Create a sphere input file using the following command:
vi INSPH
5. Paste the following into your file: ./IO86.dms R X 0.0 4.0 1.4 IO86.sph
6. Run the program with the following command sphgen -i INSPH -o OUTSPH
003: Grid/box
showbox, grid generation, visualization in Chimera
004: Minimization
Explanation of .in file for minimization and process (Chimera visuals after)
005: Rigid Docking
Explanation of .in file for rigid docking and process (Chimera visuals after)
006: Flexible Docking
Explanation of .in for flex docking (Chimera visuals after)
007: Footprint Scoring
Explanation of .in for FPS, use of Python script to generate graph
008: 5k Virtual Screen
Slurm and queue etiquette, VS .in explanation and queue submission, ViewDock in Chimera