Difference between revisions of "2025 Denovo tutorial 2 with PDBID 1XMU"
Stonybrook (talk | contribs) (→DOCK6 input) |
Stonybrook (talk | contribs) (→DOCK6 input) |
||
| Line 44: | Line 44: | ||
[[File: 1xmu_files.png|center|400px]] | [[File: 1xmu_files.png|center|400px]] | ||
| − | The 01dnv.in contains the DOCK6 input parameters. Please note that the bolded lines represent the input and output file names. | + | The 01dnv.in contains the DOCK6 input parameters. Please note that the bolded lines represent customized items for this tutorial, including the input and output file names. Additionally, we intend to modify the ligand with only one layer while keeping the original pose. DOCK6 parameter files and fragment library have been previously prepared. Please refer to the virtual screening and genetic algorithm tutorials for further instructions if needed. |
| + | |||
conformer_search_type denovo | conformer_search_type denovo | ||
| − | dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 | + | '''dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 |
dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 | dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 | ||
| − | dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2 | + | dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2''' |
dn_user_specified_anchor yes | dn_user_specified_anchor yes | ||
'''dn_fraglib_anchor_file 1xmu_lig_wDu.mol2''' | '''dn_fraglib_anchor_file 1xmu_lig_wDu.mol2''' | ||
| − | dn_torenv_table ../006_ga_lib/unique_full_sorted_fraglib.dat | + | '''dn_torenv_table ../006_ga_lib/unique_full_sorted_fraglib.dat''' |
dn_name_identifier denovo | dn_name_identifier denovo | ||
dn_sampling_method graph | dn_sampling_method graph | ||
| Line 80: | Line 81: | ||
dn_heur_matched_rmsd 2.0 | dn_heur_matched_rmsd 2.0 | ||
dn_unique_anchors 1 | dn_unique_anchors 1 | ||
| − | dn_max_grow_layers 1 | + | '''dn_max_grow_layers 1''' |
dn_max_root_size 25 | dn_max_root_size 25 | ||
dn_max_layer_size 25 | dn_max_layer_size 25 | ||
| Line 95: | Line 96: | ||
internal_energy_cutoff 100.0 | internal_energy_cutoff 100.0 | ||
use_database_filter no | use_database_filter no | ||
| − | orient_ligand no | + | '''orient_ligand no''' |
bump_filter no | bump_filter no | ||
score_molecules yes | score_molecules yes | ||
| Line 104: | Line 105: | ||
grid_score_es_scale 1 | grid_score_es_scale 1 | ||
grid_lig_efficiency no | grid_lig_efficiency no | ||
| − | grid_score_grid_prefix ../003_grid/grid | + | '''grid_score_grid_prefix ../003_grid/grid''' |
minimize_ligand yes | minimize_ligand yes | ||
minimize_anchor yes | minimize_anchor yes | ||
| Line 122: | Line 123: | ||
simplex_restraint_min no | simplex_restraint_min no | ||
atom_model all | atom_model all | ||
| − | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_AMBER_parm99.defn | + | '''vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_AMBER_parm99.defn |
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn | ||
| − | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl | + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl''' |
Revision as of 17:42, 25 February 2025
Contents
Introduction
This tutorial describes de novo method to refine ligand structure(s) using DOCK6. Rather than screening existing compounds, de novo refinement adds fragments based on known bioactive compounds, aiming to produce new ligands that bind more tightly to protein receptors than current options. This strategy is particularly valuable for:
- Improving suboptimal ligand binding profiles
- Overcoming resistance mechanisms
- Exploring novel scaffolds while maintaining critical pharmacophore features
In this tutorial, we will utilize PDB structure 1XMU - Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Roflumilast.
System Preparation
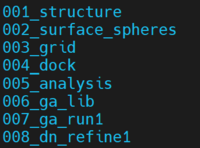
We first prepare a folder to contain all the input and output files for de novo refinement for 1XMU. This is the current directory structure under the upper directory dock6-1xmu.
Input File
Modify Ligand with Dummy Atom
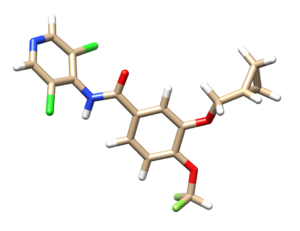
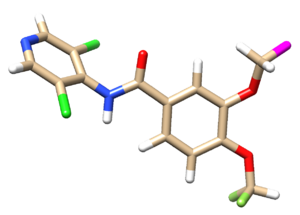
Below shows the original ligand with hydrogen (mol2 file).
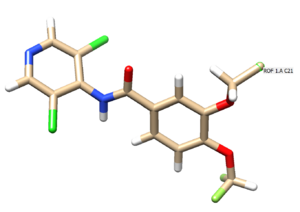
Select the atoms to delete and save the mol2 file as 1xmu_lig_wDu.mol2. Hover the cursor over the atom that you intend to change to a dummy atom. Here we are changing the C21 to a dummy atom.
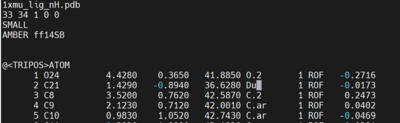
Open the 1xmu_lig_wDu.mol2 file and edit the C21 atom type to Du as shown below.
Next reopen the 1xmu_lig_wDu.mol2 file to observe if the corresponding atom is now represented as Du in magenta.
DOCK6 input
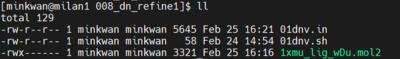
The directory 008_dn_refine1 contains the following files:
The 01dnv.in contains the DOCK6 input parameters. Please note that the bolded lines represent customized items for this tutorial, including the input and output file names. Additionally, we intend to modify the ligand with only one layer while keeping the original pose. DOCK6 parameter files and fragment library have been previously prepared. Please refer to the virtual screening and genetic algorithm tutorials for further instructions if needed.
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file 1xmu_lig_wDu.mol2 dn_torenv_table ../006_ga_lib/unique_full_sorted_fraglib.dat dn_name_identifier denovo dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_remove_duplicates yes dn_max_duplicates_per_mol 0 dn_write_pruned_duplicates no dn_advanced_pruning yes dn_prune_initial_sample yes dn_sample_torsions yes dn_prune_individual_torsions yes dn_prune_combined_torsions yes dn_random_root_selection no dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 1000 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 1 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_max_successful_att_per_root 50000 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_output_prefix 1xmu_out use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_lig_efficiency no grid_score_grid_prefix ../003_grid/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_final_min no simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl