Difference between revisions of "2025 Denovo tutorial 2 with PDBID 1XMU"
Stonybrook (talk | contribs) (→Specified Anchor Input) |
Stonybrook (talk | contribs) (→Output) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
=Introduction= | =Introduction= | ||
| − | This tutorial describes de novo method to refine ligand structure(s) using DOCK6. Rather than screening existing compounds, de novo refinement adds fragments based on known bioactive compounds, | + | This tutorial describes de novo method to refine ligand structure(s) using DOCK6. Rather than screening existing compounds, '''(1) de novo refinement''' adds fragments based on known bioactive compounds. '''(2) Generic de novo design''', on the other hand, produce new ligands from scratch. Such that we yield new ligand beyond the current available chemotypes allowing tighter binding protein receptors. This strategy is particularly valuable for: |
*Improving suboptimal ligand binding profiles | *Improving suboptimal ligand binding profiles | ||
| Line 8: | Line 8: | ||
*Exploring novel scaffolds while maintaining critical pharmacophore features | *Exploring novel scaffolds while maintaining critical pharmacophore features | ||
| − | In this tutorial, we will utilize PDB structure 1XMU - Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Roflumilast. | + | In this tutorial, we will utilize PDB structure 1XMU - Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Roflumilast. |
| − | |||
=System Preparation= | =System Preparation= | ||
| Line 18: | Line 17: | ||
=De Novo Refinement= | =De Novo Refinement= | ||
| + | |||
| + | Refinement requires users to first modify the original ligand file (mol2 with hydrogen) to specify the dummy atom location. The dummy atom is used as the starting point for the refinement process. | ||
==Modify Ligand with Dummy Atom== | ==Modify Ligand with Dummy Atom== | ||
| Line 158: | Line 159: | ||
[[File: 1xmu_output2.png|400px|center]] | [[File: 1xmu_output2.png|400px|center]] | ||
| − | |||
=De Novo Generic= | =De Novo Generic= | ||
| − | Dock6 can also perform generic de novo design without specifying the exact location to refine with the dummy atom. Below we will illustrate performing generic de novo with or without a specify anchor. This approach will allow orientation of ligand molecules. Parameters for the input file should be carefully considered, e.g. Lipinski's rule of five. | + | Dock6 can also perform generic de novo design without specifying the exact location to refine with the dummy atom. Below we will illustrate performing generic de novo '''with or without a specify anchor'''. This approach will allow orientation of ligand molecules. Parameters for the input file should be carefully considered, e.g. Lipinski's rule of five. |
| − | == | + | ==No Anchor Specification== |
In the directory '''009_dn_generic1''', we created the dock6 input file '''without the anchor specification''': | In the directory '''009_dn_generic1''', we created the dock6 input file '''without the anchor specification''': | ||
| Line 254: | Line 254: | ||
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn | ||
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl | ||
| + | |||
| + | |||
| + | We shall run this with a slurm script (01dng.slurm) similar to the one without specified anchor. | ||
| + | |||
| + | #!/bin/bash | ||
| + | # | ||
| + | #SBATCH --job-name=1xmu_dng2 | ||
| + | #SBATCH --output=generic | ||
| + | #SBATCH --ntasks=1 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --time=4:00:00 | ||
| + | #SBATCH -p ood-short-96core | ||
| + | ## at least 6 nodes for medium | ||
| + | dock6 -i 01dng.in -o 01dng.out | ||
| + | |||
| + | Submit the slurm script with: | ||
| + | |||
| + | sbatch 01dng.slurm | ||
| Line 373: | Line 391: | ||
#SBATCH -p ood-short-96core | #SBATCH -p ood-short-96core | ||
## at least 6 nodes for medium | ## at least 6 nodes for medium | ||
| − | |||
dock6 -i 01dng.in -o 01dng.out | dock6 -i 01dng.in -o 01dng.out | ||
| Line 381: | Line 398: | ||
==Output== | ==Output== | ||
| + | |||
| + | This shows the output files at the end of run for the generic de novo design with specified anchor. | ||
| + | |||
| + | [[File:1xmu_output3.png|center|400px]] | ||
| + | |||
| + | Take the last 10000 (or more) lines from the generic.completed.denovo_build.mol2 into a new mol2 file by: | ||
| + | |||
| + | tail -10000 generic.completed.denovo_build.mol2 > out.mol2 | ||
| + | |||
| + | Open the mol2 file and remove the lines until you get to the first Grid Score. For example, in our out.mol2 file for this tutorial, we need to remove 65 lines. | ||
| + | |||
| + | [[File:1xmu_output5.png|center|400px]] | ||
==Results== | ==Results== | ||
| + | |||
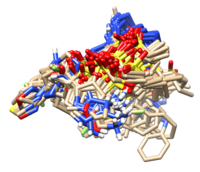
| + | Open your results in chimera and you shall see the overlapped designs. | ||
| + | |||
| + | [[File:1xmu_output4.png|center|200px]] | ||
| + | |||
| + | Again, we can analyze by using View Dock. Load the out.mol2 (''blue'') along with the 1xmu_rec_wH.mol2 (''beige''). | ||
| + | |||
| + | |||
| + | [[File:1xmu_output6.png|300px|center]] | ||
| + | |||
| + | |||
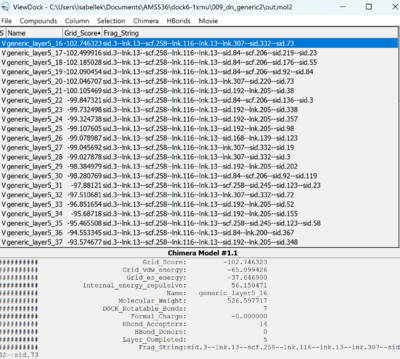
| + | We can compare the grid score and the fragment strings from the View Dock panel. | ||
| + | |||
| + | [[File:1xmu_output7.png|400px|center]] | ||
Latest revision as of 13:28, 10 March 2025
Contents
Introduction
This tutorial describes de novo method to refine ligand structure(s) using DOCK6. Rather than screening existing compounds, (1) de novo refinement adds fragments based on known bioactive compounds. (2) Generic de novo design, on the other hand, produce new ligands from scratch. Such that we yield new ligand beyond the current available chemotypes allowing tighter binding protein receptors. This strategy is particularly valuable for:
- Improving suboptimal ligand binding profiles
- Overcoming resistance mechanisms
- Exploring novel scaffolds while maintaining critical pharmacophore features
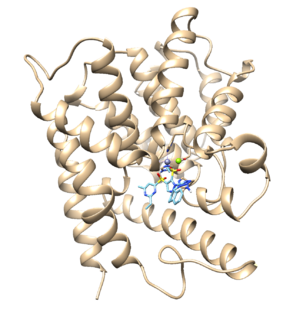
In this tutorial, we will utilize PDB structure 1XMU - Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Roflumilast.
System Preparation
We first prepare a folder to contain all the input and output files for de novo refinement for 1XMU. This is the current directory structure under the upper directory dock6-1xmu.
De Novo Refinement
Refinement requires users to first modify the original ligand file (mol2 with hydrogen) to specify the dummy atom location. The dummy atom is used as the starting point for the refinement process.
Modify Ligand with Dummy Atom
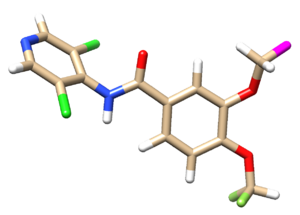
Below shows the original ligand with hydrogen (mol2 file).
Select the atoms to delete and save the mol2 file as 1xmu_lig_wDu.mol2. Hover the cursor over the atom that you intend to change to a dummy atom. Here we are changing the C21 to a dummy atom.
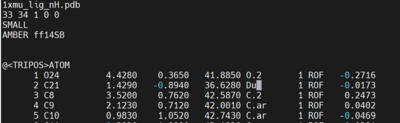
Open the 1xmu_lig_wDu.mol2 file and edit the C21 atom type to Du as shown below.
Next reopen the 1xmu_lig_wDu.mol2 file to observe if the corresponding atom is now represented as Du in magenta.
DOCK6 input
The directory 008_dn_refine1 contains the following files:
The 01dnv.in contains the DOCK6 input parameters. Please note that the bolded lines represent customized items for this tutorial, including the input and output file names. Additionally, we intend to modify the ligand with only one layer while keeping the original pose. DOCK6 parameter files and fragment libraries have been previously prepared. Please refer to the virtual screening and genetic algorithm tutorials for further instructions if needed.
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file 1xmu_lig_wDu.mol2 dn_torenv_table ../006_ga_lib/unique_full_sorted_fraglib.dat dn_name_identifier denovo dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_remove_duplicates yes dn_max_duplicates_per_mol 0 dn_write_pruned_duplicates no dn_advanced_pruning yes dn_prune_initial_sample yes dn_sample_torsions yes dn_prune_individual_torsions yes dn_prune_combined_torsions yes dn_random_root_selection no dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 1000 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 1 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_max_successful_att_per_root 50000 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_output_prefix 1xmu_out use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_lig_efficiency no grid_score_grid_prefix ../003_grid/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_final_min no simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl
Then, run the bash script, which contains:
#!/bin/bash dock6 -i 01dnv.in -o 01dnv.out >&01dnv.log&
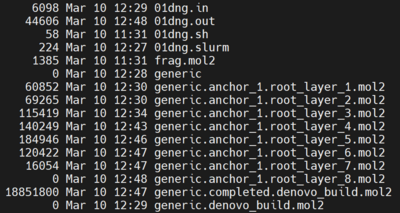
Output Files
When completed, the directory now contains:
1xmu_out.denovo_build.mol2 contains no data, which is an intermediate file genereated.
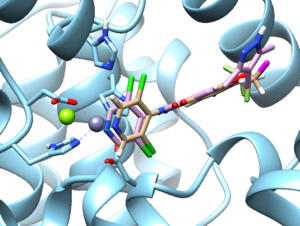
Results
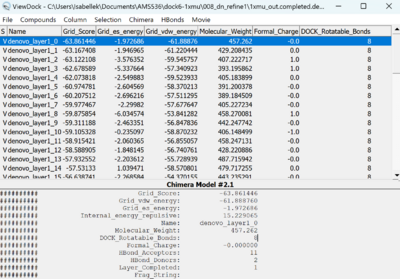
From ChimeraX > Tools> Structure/Binding Analysis > View Dock, load the 1xmu_out.completed.denovo_build.mol2 (pink) along with the 1xmu_rec_wH.mol2 (blue) and 1xmu_lig_wH.mol2 (beige). We can see the refinement at the dummy atom (magenta)
We can compare the interaction energy from the View Dock panel.
De Novo Generic
Dock6 can also perform generic de novo design without specifying the exact location to refine with the dummy atom. Below we will illustrate performing generic de novo with or without a specify anchor. This approach will allow orientation of ligand molecules. Parameters for the input file should be carefully considered, e.g. Lipinski's rule of five.
No Anchor Specification
In the directory 009_dn_generic1, we created the dock6 input file without the anchor specification:
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_torenv.dat dn_name_identifier generic dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_remove_duplicates yes dn_max_duplicates_per_mol 0 dn_write_pruned_duplicates no dn_advanced_pruning yes dn_prune_initial_sample yes dn_sample_torsions yes dn_prune_individual_torsions yes dn_prune_combined_torsions yes dn_random_root_selection no dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_max_successful_att_per_root 50000 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_write_growth_trees no dn_output_prefix generic use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002_surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_lig_efficiency no grid_score_grid_prefix ../003_grid/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_final_min no simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl
We shall run this with a slurm script (01dng.slurm) similar to the one without specified anchor.
#!/bin/bash # #SBATCH --job-name=1xmu_dng2 #SBATCH --output=generic #SBATCH --ntasks=1 #SBATCH --nodes=1 #SBATCH --time=4:00:00 #SBATCH -p ood-short-96core ## at least 6 nodes for medium dock6 -i 01dng.in -o 01dng.out
Submit the slurm script with:
sbatch 01dng.slurm
Specified Anchor Input
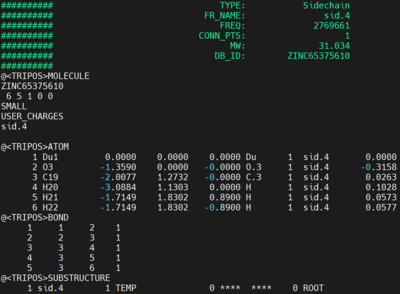
We first need to create the mol2 file containing the fragment to use as anchor. You can look over the list of fragments (scaffold/linker/side chains) in the following file:
/gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2
When you identify the fragment of your preference, you can copy or cat into a new mol2 file. File name frag.mol2 is used in this tutorial. The copied content should contain the following:
In the directory 009_dn_generic2, we created the dock6 input file with the anchor specification:
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file frag.mol2 dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/fraglib_torenv.dat dn_name_identifier generic dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_remove_duplicates yes dn_max_duplicates_per_mol 0 dn_write_pruned_duplicates no dn_advanced_pruning yes dn_prune_initial_sample yes dn_sample_torsions yes dn_prune_individual_torsions yes dn_prune_combined_torsions yes dn_random_root_selection no dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_max_successful_att_per_root 50000 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_write_growth_trees no dn_output_prefix generic use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002_surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_lig_efficiency no grid_score_grid_prefix ../003_grid/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_final_min no simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.12_ams536/parameters/flex_drive.tbl
We shall run this with a slurm script (01dng.slurm) similar to the one without specified anchor.
#!/bin/bash # #SBATCH --job-name=1xmu_dng2 #SBATCH --output=generic #SBATCH --ntasks=1 #SBATCH --nodes=1 #SBATCH --time=4:00:00 #SBATCH -p ood-short-96core ## at least 6 nodes for medium dock6 -i 01dng.in -o 01dng.out
Submit the slurm script with:
sbatch 01dng.slurm
Output
This shows the output files at the end of run for the generic de novo design with specified anchor.
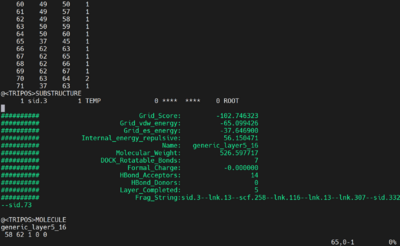
Take the last 10000 (or more) lines from the generic.completed.denovo_build.mol2 into a new mol2 file by:
tail -10000 generic.completed.denovo_build.mol2 > out.mol2
Open the mol2 file and remove the lines until you get to the first Grid Score. For example, in our out.mol2 file for this tutorial, we need to remove 65 lines.
Results
Open your results in chimera and you shall see the overlapped designs.
Again, we can analyze by using View Dock. Load the out.mol2 (blue) along with the 1xmu_rec_wH.mol2 (beige).
We can compare the grid score and the fragment strings from the View Dock panel.