Difference between revisions of "2013 DOCK tutorial with Orotodine Monophosphate Decarboxylase"
(Created page with "For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link [http://www.unilang.org/wiki/index.php/Wiki_markup Wiki Markup] as a reference for editing the wiki. T...") |
Stonybrook (talk | contribs) |
||
| (144 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | For additional Rizzo Lab tutorials see [[DOCK Tutorials]]. Use this link [http://www. | + | For additional Rizzo Lab tutorials see [[DOCK Tutorials]]. Use this link [http://www.mediawiki.org/wiki/Help:Formatting Wiki Formatting] as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2013, using DOCK v6.6. It is assumed that users of this tutorial have a basic understanding of the command line and are able to navigate a Unix operating system. |
==I. Introduction== | ==I. Introduction== | ||
| Line 13: | Line 13: | ||
# The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations. | # The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations. | ||
| − | + | ===Orotodine Monophosphate Decarboxylase=== | |
| − | === | + | The protein receptor which is the subject of this tutorial is orotodine monophosphate decarboxylase (OMP), a homodimeric protein from the organism ''Methanobacterium thermoautotrophicum''. OMP is involved in the biosynthesis of several pyrimidines including uridine monophosphate (UMP), the ligand used in this tutorial. UMP is a pyrimidine-based nucleotide monomer of RNA. The structure used for this tutorial can be found at the Protein Data Bank under accenssion number [http://www.pdb.org/pdb/explore/explore.do?structureId=1LOQ 1LOQ]. |
| − | |||
| − | |||
| − | |||
| − | ''' | ||
| − | is | ||
| − | |||
===Organizing Directories=== | ===Organizing Directories=== | ||
| − | While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find | + | While performing docking, it is convenient to create a project directory (here we call it ~AMS536/dock-tutorial/), and adopt a standard directory structure / naming scheme, so that files are easy to find and identify. For this tutorial, we will use something similar to the following: |
| − | ~username/AMS536/ | + | ~username/AMS536/dock-tutorial/00.files/ |
| − | /01 | + | /01.dockprep/ |
| − | /02 | + | /02.surface-sphgen/ |
| − | /03 | + | /03.box-grid/ |
| − | /04 | + | /04.dock/ |
| − | /05-virtual-screen/ | + | /05.mini-virtual-screen/ |
| − | + | /06.database-filter/ | |
| − | In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, ' | + | /07.virtual-screen/ |
| + | |||
| + | Before beginning the tutorial, it may be easiest to make all of the sub-directories at the same time. From here on in, these subdirectories will be referred to as ~00.files/, ~01.dockprep/ etc. In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '1LOQ'. | ||
==II. Preparing the Receptor and Ligand== | ==II. Preparing the Receptor and Ligand== | ||
| − | ===Downloading the PDB | + | ===Downloading the PDB file (1LOQ)=== |
| − | + | Go to PDB homepage (http://www.rcsb.org/pdb/home/home.do ) enter the protein ID (1LOQ) in the search bar, click Download Files in the top-right of the webpage, then select PDB File (text). In the new window, save the file in your ~00.files/ folder. | |
| − | ===Preparing the | + | ===Preparing the ligand and receptor in Chimera=== |
| − | If you are | + | NOTE: If you are accessing files remotely (i.e. through a terminal) and want to use a visualization program such as Chimera, first transfer the files to your local computer, run Chimera, and transfer the files back. |
| − | + | In this section, we will create four new files and save them in the ~01.dockprep/ folder: | |
| − | + | 1LOQ.dockprep.mol2 (complete system with hydrogens and charges) | |
| + | 1LOQ.receptor.mol2 (the receptor alone) | ||
| + | 1LOQ.receptor.noH.pdb (the receptor alone, without hydrogens) | ||
| + | 1LOQ.ligand.mol2 (the ligand alone) | ||
| − | + | To prepare these files, first make a copy of the original PDB file in the ~00.files/ folder (named, for example, 1LOQ.copy.pdb) and open it with VIM ($ vim 1LOQ.copy.pdb). Because the residue name of the ligand (U) will give us some problems when assigning charges, change the residue name "U" to "LIG" starting at line 2082. Here is an example command that will change all instances of " U" into "LIG", while preserving the correct spacing. Note there are two blank spaces before U in the following expression: | |
| − | + | %s/ U/LIG/gc | |
| − | |||
| − | + | For this command, '''g''' is short for global and '''c''' is short for check with the user before making the change. | |
| − | + | Next, open up the PDB file (1LOQ.copy.pdb) in Chimera. To delete water molecules and other ligands, click ''Tools'' -> ''Structure Editing'' -> ''Dock Prep''. Check all boxes and click ''Okay'' to the end. Alternatively, the waters can be deleted manually by choosing ''Select'' -> ''Residue'' -> ''HOH'', then go to ''Actions'' -> ''Atoms/Bonds'' -> ''Delete''. Hydrogen atoms can be added manually by choosing ''Tools'' -> ''Structure Editing'' -> ''Add H''. | |
| − | + | Next, to add charges to the ligand and receptor, go to ''Select'' -> ''Residue'' -> ''LIG'', then go to ''Tools'' -> ''Structure Editing'' -> ''Add Charge''. Choose ''AMBER ff99SB'' as the charge model, click ''Okay'', and when prompted chose AM1-BCC charges for the ligand, and make sure the ''Net Charge'' is set to ''-1''. (You must consider the chemistry of the ligand when assigning a charge state). | |
| − | + | Finally, save this file as '''1LOQ.dockprep.mol2''' into your ~01.dockprep/ folder. | |
| − | + | ===Generating the final files=== | |
| − | + | To create the receptor file: Open 1LOQ.dockprep.mol2, click ''Select'' -> ''Residue'' -> ''LIG''. Then click ''Actions'' -> ''Atoms/Bonds'' -> ''Delete''. Save the file as '''1LOQ.receptor.mol2'''. | |
| − | + | To create the receptor file with no hydrogen atoms: Open 1LOQ.receptor.mol2, click ''Select'' -> ''Chemistry'' -> ''Element'' -> ''H'', then chose ''Actions'' -> ''Atoms/Bonds'' -> ''Delete''. Save the file as '''1LOQ.receptor.noH.pdb'''. | |
| − | Save the file as | + | To create the ligand file: Open 1LOQ.dockprep.mol2, click ''Select'' -> ''Residue'' -> ''LIG'', then click ''Select'' -> ''Invert'', then chose ''Actions'' -> ''Atoms/Bonds'' -> ''Delete''. Save the file as '''1LOQ.ligand.mol2'''. |
| − | + | ==III. Generating Receptor Surface and Spheres== | |
| − | + | === Receptor Surface === | |
| − | + | The next section of this tutorial will be performed in your ~02.surface-sphgen/ folder. | |
| − | + | In order to generate the receptor surface one can perform the following steps (working in Chimera): | |
| + | # Go ''File -> Open'' and choose the PDB file of the protein containing no hydrogens (1LOQ.receptor.noH.pdb) from '''01.dockprep''' | ||
| + | # Further, ''Actions'' -> ''Surface'' -> ''Show'' | ||
| + | # Go ''Tools'' -> ''Structure Editing'' -> ''Write DMS'' in order to obtain a ''dms'' file, which we will need while placing spheres | ||
| + | # In the new window save the surface as '''1LOQ.receptor.dms''' | ||
| − | |||
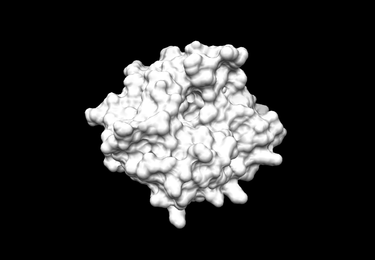
| − | + | [[Image:Receptor surface.png|thumb|center|375px|Receptor surface]] | |
| − | + | === Placing Spheres === | |
| − | + | For more information SPHGEN, please consult the DOCK online owners manual at: | |
| − | + | http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm | |
| + | In order to place the spheres on the receptor surface, the following steps are required: | ||
| − | + | '''1.''' Create a file called '''INSPH''' (an input file we will need later) and fill it out as follows (parameter descriptions included to the right), then save it. | |
| − | + | 1LOQ.receptor.dms | |
| + | R | ||
| + | X | ||
| + | 0.0 | ||
| + | 4.0 | ||
| + | 1.4 | ||
| + | 1LOQ.receptor.sph | ||
| − | + | Here is a line-by-line break down on what each line does: | |
| − | + | *1LOQ.receptor.dms - molecular surface file that we got from previous step | |
| + | *R - sphere outside of surface (R) or inside surface (L) | ||
| + | *X - specifies subset of surface points to be used (X=all points) | ||
| + | *0.0 - prevents generation of large spheres with close surface contacts(defalut=0.0) | ||
| + | *4.0 - maximum sphere radius in angstroms (default=4.0) | ||
| + | *1.4 - minimum sphere radius in angstroms (default=radius of probe) | ||
| + | *1LOQ.receptor.sph - clustered spheres file that we want to generate | ||
| − | + | '''2.''' Run the sphgen program from the command line: | |
| − | + | sphgen -i INSPH -o OUTSPH | |
| − | + | '''OUTSPH''' is the output file containing the information on the sphgen run, while '''1LOQ.receptor.sph''' is the file which contains the spheres. | |
| − | + | '''3.''' ''(optional)'' If one wants to have a look at the spheres received as an outcome of sphgen, one should run '''showsphere''', by typing in the command line: | |
| − | + | showsphere | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The following questions will be prompted on the command line: | |
| − | + | Enter name of sphere cluster file: | |
| + | Enter cluster number to process (<0 = all): | ||
| + | Generate surfaces as well as pdb files (<N>/Y)? | ||
| + | Enter name for output file prefix: | ||
| + | Process cluster 0 (contains ALL spheres) (<N>/Y)? | ||
| − | + | As the questions have been answered the program will generate the spheres, writing the data in a '''PDB''' file, afterwards the spheres may be observed by opening this file in '''Chimera'''. | |
| − | + | '''4.''' Run the '''sphere_selector''' program from the command line as follows: | |
| − | |||
| − | + | sphere_selector 1LOQ.receptor.sph ../01.dockprep/1LOQ.ligand.mol2 8.0 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | This program selects the spheres within a user-defined radius (8.0 here) of a target molecule from the previously obtained file '''1LOQ.receptor.sph'''. As a result a new file '''selected_spheres.sph''' will be generated. Again, the spheres can and should be visualized by using '''showsphere''' and '''Chimera'''. | |
| − | + | '''5.''' Run '''showsphere''' in order to visualize the spheres: | |
showsphere | showsphere | ||
| − | + | When prompted on the command line, answering the questions as follows: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | selected_spheres.sph | |
-1 | -1 | ||
N | N | ||
| − | + | output_spheres_selected | |
N | N | ||
| − | + | * Launch '''Chimera''', choose ''File'' -> ''Open'', choose '''1LOQ.receptor.noH.pdb''' | |
| + | * Go ''File'' -> ''Open'', choosing '''output_spheres_selected.pdb''' | ||
| + | * Go ''Select'' -> ''Residue'' -> ''SPH'' | ||
| + | * Finally, ''Actions'' -> ''Atoms/Bonds'' -> ''sphere'' | ||
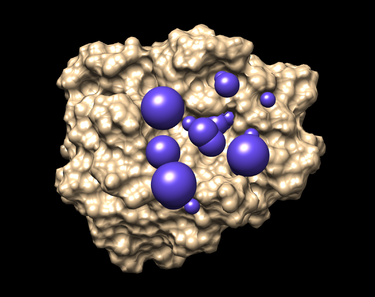
| − | + | The picture like one placed below will be obtained afterwards. | |
| − | + | [[Image:Selected spheres.png|thumb|375px|center|Selected spheres]] | |
| − | + | ==IV. Generating Box and Grid== | |
| − | + | This section of the tutorial will be performed in your ~03.box-grid/ folder. For more information GRID, please consult the DOCK online owners manual at: | |
| − | + | http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm | |
| − | + | ===Box=== | |
| − | + | We will create a box around the binding site to define the boundaries of ligand sampling and aid in energy calculations. As a general rule, we will create a box around the spheres (from the previous step) with margins of at least 8.0 Angstroms on all sides. You first need to create a '''showbox.in''' file to generate the box: | |
| − | + | vim showbox.in | |
| − | + | Edit this file as following: | |
| − | + | Y # Yes, generate a box | |
| − | + | 8.0 # box margin | |
| + | ../02.surface-sphgen/selected_spheres.sph # input sphere files | ||
| + | 1 # which sphere cluster to use | ||
| + | 1LOQ.box.pdb # output file name | ||
| + | |||
| + | ''Note: Do not include the comments in the showbox.in file''. To run the program '''showbox''', give the name of the program on the command line and provide the '''showbox.in''' file: | ||
| − | + | showbox < showbox.in | |
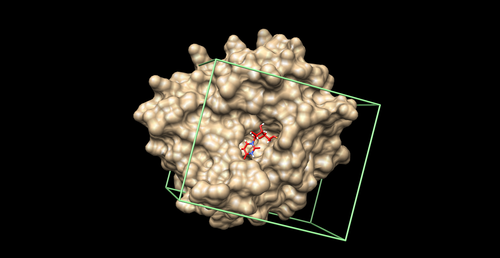
| − | + | [[Image:boxexample.png|thumb|500px|center|Box Image]] | |
| − | |||
| − | |||
| − | + | ===Grid=== | |
| − | |||
| − | |||
| − | |||
| − | + | Grids are used as a method to speed up evaluation of the energy scoring function by pre-computing electrostatic and VDW potential energies on the receptor at defined grid points within the box. VDW parameters are stored in a file that was installed with DOCK, and we will find it convenient to copy it and two other files to our ~00.files/ folder. In this class, DOCK was installed in the following location: | |
| − | + | /home/wjallen/AMS536/local/dock6/ | |
| − | + | You can determine the path to your DOCK installation by typing: | |
| − | + | which dock6 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | From that location, copy the following three files to your local ~00.files/ folder: | |
| − | |||
| − | + | /home/wjallen/AMS536/local/dock6/parameters/vdw_AMBER_parm99.def | |
| − | + | /home/wjallen/AMS536/local/dock6/parameters/flex.defn | |
| + | /home/wjallen/AMS536/local/dock6/parameters/flex_drive.tbl | ||
| − | + | Use grid program to generate the grid and answer the questions as following: | |
| − | + | touch grid.in | |
| − | + | grid -i grid.in | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
compute_grids yes | compute_grids yes | ||
| − | grid_spacing 0. | + | grid_spacing 0.4 |
output_molecule no | output_molecule no | ||
contact_score no | contact_score no | ||
| Line 240: | Line 219: | ||
bump_filter yes | bump_filter yes | ||
bump_overlap 0.75 | bump_overlap 0.75 | ||
| − | receptor_file ../01 | + | receptor_file ../01.dockprep/1LOQ.receptor.mol2 |
| − | box_file | + | box_file ../03.box-grid/1LOQ.box.pdb |
| − | vdw_definition_file / | + | vdw_definition_file ../00.files/vdw_AMBER_parm99.defn |
| − | score_grid_prefix grid | + | score_grid_prefix 1LOQ.grid |
| − | |||
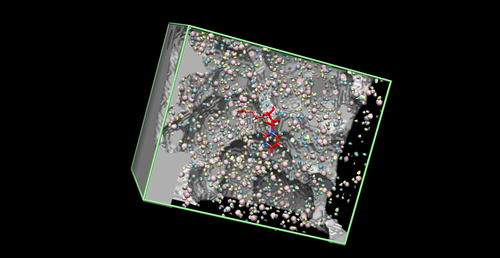
| − | + | [[Image:gridexample.png|thumb|500px|center|Grid Image]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==V. Docking a Single Molecule for Pose Reproduction== | ==V. Docking a Single Molecule for Pose Reproduction== | ||
| − | + | This part of the tutorial will be performed in your ~04.dock/ folder. For more information DOCK, please consult the DOCK online owners manual at: | |
| − | |||
| − | + | http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm | |
| − | + | ===Rigid Docking=== | |
| − | + | When docking a molecule ''rigidly'' in DOCK, no conformations or rotamers will be sampled other than the input configuration which, in our case, was derived from the crystal structure. This will be much faster that ''flexible'' docking, but it is not appropriate for virtual screening because the docked conformation of compounds from a library is generally not known. First create an empty input file for docking: | |
| − | + | touch dock.in | |
| − | + | Then execute DOCK6: | |
| − | + | dock6 -i dock.in | |
| − | + | You will be prompted for answers to many questions. For the rigid docking experiments, suggested parameters are as follows: | |
| − | + | ligand_atom_file ../01.dockprep/1LOQ.ligand.mol2 | |
| − | + | limit_max_ligands no | |
| − | + | skip_molecule no | |
| − | + | read_mol_solvation no | |
| − | + | calculate_rmsd no | |
| − | + | use_database_filter no | |
| − | + | orient_ligand yes | |
| − | + | automated_matching yes | |
| − | + | receptor_site_file ../02.surface-sphgen/selected_spheres.sph | |
| − | ligand_atom_file | + | use_internal_energy no |
| − | + | flexible_ligand no | |
| − | + | bump_filter no | |
| − | + | score_molecules yes | |
| − | + | contact_score_primary no | |
| − | + | contact_score_secondary no | |
| − | + | grid_score_primary yes | |
| − | + | grid_score_secondary no | |
| − | + | grid_score_rep_rad_scale 1 | |
| − | + | grid_score_vdw_scale 1 | |
| − | + | grid_score_es_scale 1 | |
| − | + | grid_score_grid_prefix ../03.box-grid/1LOQ.grid | |
| − | + | multigrid_score_secondary no | |
| − | + | dock3.5_score_secondary no | |
| − | + | continuous_score_secondary no | |
| − | + | descriptor_score_secondary no | |
| − | + | gbsa_zou_score_secondary no | |
| − | + | gbsa_hawkins_score_secondary no | |
| − | vdw_defn_file | + | SASA_descriptor_score_secondary no |
| − | flex_defn_file | + | amber_score_secondary no |
| − | flex_drive_file | + | minimize_ligand no |
| − | + | atom_model all | |
| − | + | vdw_defn_file ../00.files/vdw_AMBER_parm99.defn | |
| + | flex_defn_file ../00.files/flex.defn | ||
| + | flex_drive_file ../00.files/flex_drive.tbl | ||
| + | ligand_outfile_prefix output | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| − | + | With these input parameters, the rigid docking experiment will result in a single scored ligand pose. Open Chimera to visualize the docking result. After opening the receptor file in surface view, open the ligand file (named "output_scored.mol2") by going under ''Tools'' -> ''Surface/Binding Analysis'' -> ''ViewDock''. | |
| − | + | [[Image:dock_original ligand.png|thumb|375px|center|Rigid docking result]] | |
| − | + | ===Flexible Docking=== | |
| − | + | Flexible docking will take longer, but is necessary for virtual screening. For flexible docking, several input parameters need to be changed, and these changes will generate additional questions prompted to the user. Some changes to parameters are shown below: | |
| − | |||
| − | |||
| − | |||
| − | |||
calculate_rmsd yes | calculate_rmsd yes | ||
| − | + | flexible_ligand yes | |
| + | use_internal_energy yes | ||
| + | minimize_ligand yes | ||
num_scored_conformers 10 | num_scored_conformers 10 | ||
| − | + | Once these changes have been made to the input file, execute DOCK6 again: | |
| + | dock6 -i dock.in | ||
| − | + | This flexible ligand protocol can be used with multiple ligand input files. The docked ligand poses can be compared using RMSD (Root Mean Squared Deviation). RMSD is a measurement of the distance between atoms on the original crystal structure ligand with the newly generated ligand conformation. An RMSD under 2 Angstroms would indicate a good conformation. Below are two examples of a final docked ligand in the receptor binding site pocket. The worst and best energy scored ligands are shown below. The original ligand is in cyan and the docked ligand is colored in heteroatom. | |
| − | + | [[Image:dock_bestligand_and original.png|thumb|375px|center|Best scored ligand result (-78.3 kcal/mol)]] | |
| − | + | [[Image:dock_worstligand_and original.png|thumb|375px|center|Worst scored ligand result (-67.4 kcal/mol)]] | |
| − | |||
| − | |||
| − | + | ==VI. Virtual Screening== | |
| − | + | ===Virtual Screening Introduction=== | |
| − | + | A virtual screen of various ligand allows for the comparison of both qualitative (e.g. position in binding site) and quantitative (e.g. energy scores) data pertaining to the each screened ligand with an originally docked molecule. Virtual screening is often used as a method to cut the cost of experimentation by narrowing down the ligands within a database and predicting which will exhibit the most favorable binding to a specific protein (with a pre-determined .grid file). | |
| − | + | ===Running the Database Filter=== | |
| − | + | When first beginning the virtual screen, a filter should be specified in order save computational power by only screening potentially effective ligands within the directory ~06.database-filter/. This involves limiting several variables of ligands specified in a .mol2 file. To limit the ligands from the original database, a file "dock_filter.in" is created and executed using DOCK. Note that in this example the file "cdiv_p0.0.mol2" is a multi mol2 file containing thousands of ligands from the CHEMDIV vendor which was previously downloaded from the UCSF ZINC database (https://zinc.docking.org/about/ucsf). ZINC is a great resource of purchasable compounds for virtual screening. Here are the contents of the file: | |
| − | + | ligand_atom_file /home/wjallen/AMS536/multi-mol2-files/cdiv_p0.0.mol2 | |
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter yes | ||
| + | dbfilter_max_heavy_atoms 50 | ||
| + | dbfilter_min_heavy_atoms 0 | ||
| + | dbfilter_max_rot_bonds 10 | ||
| + | dbfilter_min_rot_bonds 0 | ||
| + | dbfilter_max_molwt 9999.0 | ||
| + | dbfilter_min_molwt 0.0 | ||
| + | dbfilter_max_formal_charge -1.0 | ||
| + | dbfilter_min_formal_charge -2.0 | ||
| + | orient_ligand no | ||
| + | use_internal_energy no | ||
| + | flexible_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules no | ||
| + | atom_model all | ||
| + | vdw_defn_file ../00.files/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file ../00.files/flex.defn | ||
| + | flex_drive_file ../00.files/flex_drive.tbl | ||
| + | ligand_outfile_prefix filtered | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| − | + | In this file some important characteristics of the ligands that are limited are ''dbfilter_max_heavy_atoms'', ''dbfilter_max_rot_bonds'', ''dbfilter_max_molwt'', and both ''dbfilter_max_formal_charge'' and ''dbfilter_min_formal_charge''. As show in the text of the file, a file called filtered_scored.mol2 will be created with all of the ligands from the original database that fit into the specified limits of number of heavy atoms, number of rotatable bonds, molecular weight, and formal charge. '''''Note:''''' ''The max and min formal charge specified in this file (-1 to -2) are an extremely limited range, and are only used in this tutorial as an illustration.'' | |
| − | + | ===Running the Virtual Screen=== | |
| − | |||
| − | + | After creating a new directory, ~07.virtual-screen/, a dock.in file must be created. So as not to confuse the virtual screen file with the original dock.in of the docked ligand, the new file created is called dock_vs.in. Running this file in dock6 allows us to fill in the important details of the file that terminate in a virtual screening of the input ligand. | |
| − | + | ligand_atom_file ../06.database-filter/filtered_scored.mol2 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
limit_max_ligands no | limit_max_ligands no | ||
skip_molecule no | skip_molecule no | ||
read_mol_solvation no | read_mol_solvation no | ||
| − | calculate_rmsd | + | calculate_rmsd no |
| − | |||
use_database_filter no | use_database_filter no | ||
orient_ligand yes | orient_ligand yes | ||
automated_matching yes | automated_matching yes | ||
| − | receptor_site_file ../02 | + | receptor_site_file ../02.surface-sphgen/selected_spheres.sph |
| − | max_orientations | + | max_orientations 1000 |
critical_points no | critical_points no | ||
chemical_matching no | chemical_matching no | ||
| Line 398: | Line 361: | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
flexible_ligand yes | flexible_ligand yes | ||
| − | min_anchor_size | + | user_specified_anchor no |
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
pruning_use_clustering yes | pruning_use_clustering yes | ||
| − | pruning_max_orients | + | pruning_max_orients 1000 |
pruning_clustering_cutoff 100 | pruning_clustering_cutoff 100 | ||
| − | pruning_conformer_score_cutoff | + | pruning_conformer_score_cutoff 100 |
use_clash_overlap no | use_clash_overlap no | ||
write_growth_tree no | write_growth_tree no | ||
| Line 414: | Line 379: | ||
grid_score_vdw_scale 1 | grid_score_vdw_scale 1 | ||
grid_score_es_scale 1 | grid_score_es_scale 1 | ||
| − | grid_score_grid_prefix ../03-grid/ | + | grid_score_grid_prefix ../03.box-grid/1LOQ.grid |
| + | multigrid_score_secondary no | ||
dock3.5_score_secondary no | dock3.5_score_secondary no | ||
continuous_score_secondary no | continuous_score_secondary no | ||
| + | descriptor_score_secondary no | ||
gbsa_zou_score_secondary no | gbsa_zou_score_secondary no | ||
gbsa_hawkins_score_secondary no | gbsa_hawkins_score_secondary no | ||
| + | SASA_descriptor_score_secondary no | ||
amber_score_secondary no | amber_score_secondary no | ||
minimize_ligand yes | minimize_ligand yes | ||
| Line 436: | Line 404: | ||
simplex_restraint_min no | simplex_restraint_min no | ||
atom_model all | atom_model all | ||
| − | vdw_defn_file ../ | + | vdw_defn_file ../00.files/vdw_AMBER_parm99.defn |
| − | flex_defn_file ../ | + | flex_defn_file ../00.files/flex.defn |
| − | flex_drive_file ../ | + | flex_drive_file ../00.files/flex_drive.tbl |
| − | ligand_outfile_prefix | + | ligand_outfile_prefix output |
write_orientations no | write_orientations no | ||
| − | num_scored_conformers | + | num_scored_conformers 1 |
| − | |||
| − | |||
| − | |||
rank_ligands no | rank_ligands no | ||
| − | |||
| − | |||
| − | + | A few of the questions addressed in the terminal window while running a virtual screen are as follows: | |
| − | + | '''ligand_atom_file''' - here we have specified the new file created after filtering the initial ligand database file | |
| − | + | '''calculate_rmsd''' - we select the default <no> because since we are screening a library of compounds, there will be no reference position to compare them to | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | '''orient_ligand''' - here we would like to the have the ligand oriented in the binding pocket | |
| − | + | '''receptor_site_file''' - this is our selected_spheres.sph file from earlier | |
| − | + | '''internal_energy_rep_exp''' - here we select 12 rather than 9 because using a repulsive exponent of 12 gives us a slightly steeper rising potential as radius between the atoms falls below the optimal distance (sigma) | |
| − | + | '''flexible_ligand''' - when performing a virtual screen of a random database it is always advisable to set the ligands to flexible | |
| − | + | '''min_anchor_size''' - ensures that major features on the molecules are used as anchors and not just simple small groups | |
| − | + | '''score_molecules''' - we always want to score the molecules of a virtual screen in order to determine which are the best fit for the binding pocket | |
| − | + | '''grid_score_primary''' - we want the grid score to be the primary method of scoring the screened ligands | |
| − | |||
| − | + | '''grid_score_grid_prefix''' - the input for the .grid file created earlier | |
| − | |||
| − | |||
| − | |||
| − | + | '''atom_model''' - we select the all atom model | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | '''num_scored_conformers''' - we only want to produce one scored conformer from each screened ligand because we only want the best conformations | |
| − | |||
| − | + | Note that if you are performing a small sample virtual screening it is possible to run it on a local computer in a reasonable amount of time; however, if you are running the virtual screening to a very large scale (i.e. hundreds or thousands of ligands) you will need to use several nodes in Seawulf to produce results in a timely fashion. | |
| − | + | ===Analyzing the Results=== | |
| − | |||
| − | |||
| − | |||
| − | + | When your virtual screening has run completely and you are ready to analyze the results, open Chimera. In Chimera you can prepare your protein by opening 1LOQ.receptor.noH.pdb and choosing ''Actions'' -> ''Surface'' -> ''show''. You may also wish to open up the original ligand file 1LOQ.ligand.mol2 in order to show the position of the original ligand/substrate in the receptor's active site. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | To view the results of your virtual screening go to ''Tools'' -> ''Surface/Binding Analysis'' -> ''ViewDock'' and from their you can select your virtual screen output file. In the "ViewDock" pop-up window you can then select ''Column'' --> ''Show'' --> ''Grid Score'' which will add a column to the ViewDock window with the grid score of each ligand. You can double-click on the column heading ''Grid Score'' to sort the ligands based on their grid scores. | |
| − | |||
| − | + | [Chimera image1] [Chimera image2] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | The | + | ==VII. Running DOCK in Serial and in Parallel on Seawulf== |
| − | + | The [http://www.stonybrook.edu/seawulfcluster/ Seawulf] Cluster is a 470-processor Linux Cluster capable of highly parallel processing. This parallel processing allows dock virtual screens to be completed in a fraction of the time as a single processor. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | If you | + | If you are docking multiple ligands, you can use more than one processor in parallel mode, but you should never use more processors than you have ligands. Before we can run DOCK on Seawulf, we need to copy the proper files from Herbie to Seawulf. If we CD into the AMS536 folder we can use the following command from the mathlab computer to copy all of the dock-tutorial files |
| − | |||
| − | + | scp -r /dock-tutorial/ username@herbie.mathlab.sunysb.edu:~/AMS536/ | |
| − | + | Now we have all of our DOCK preparation files and folders on the seawulf cluster. | |
| − | == | + | ===Running DOCK in Serial on a Single Processor=== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Running on a single processor is very similar to running dock on the mathlab computer. | |
| − | + | If you make a file called qsub.csh with the text: | |
#!/bin/tcsh | #!/bin/tcsh | ||
| Line 586: | Line 472: | ||
#PBS -o pbs.out | #PBS -o pbs.out | ||
| − | cd /nfs/user03/ | + | cd /nfs/user03/zfoda/AMS536/dock-tutorial/07.virtscreen |
| − | /nfs/user03/ | + | /nfs/user03/wjallen/local/dock6/bin/dock6 -i dock.in -o dock.out |
| − | + | An explanation of the commands: | |
#!/bin/tcsh #Execute script with tcsh | #!/bin/tcsh #Execute script with tcsh | ||
#PBS -l nodes=1:ppn=1 #Use one node, and one processor per node, so one single processor | #PBS -l nodes=1:ppn=1 #Use one node, and one processor per node, so one single processor | ||
| − | #PBS -l walltime=01:00:00 #Allow 1 hour for your job run | + | #PBS -l walltime=01:00:00 #Allow 1 hour for your job run |
#PBS -N dock6 #Name of your job | #PBS -N dock6 #Name of your job | ||
#PBS -M user@ic.sunysb.edu #Get an email notifying you when your job is completed | #PBS -M user@ic.sunysb.edu #Get an email notifying you when your job is completed | ||
| Line 599: | Line 485: | ||
#PBS -o pbs.out #Name of your output file | #PBS -o pbs.out #Name of your output file | ||
| − | cd / | + | cd /nfs/user03/zfoda/AMS536/dock-tutorial/07.virtscreen #Change to your home directory and folder with dock files |
| − | /nfs/user03/ | + | /nfs/user03/wjallen/local/dock6/bin/dock6 -i dock.in -o dock.out #Specifies path to dock executable and provide input and output filenames |
| − | + | To submit the experiment use the command: | |
| − | + | qsub qsub.csh | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | You will have submitted a DOCK experiment to the seawulf queue. | |
| − | + | See also [http://ringo.ams.sunysb.edu/index.php/PBS_Queue PBS] commands. | |
| − | === | + | ===Running DOCK in Parallel using MPI=== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | In order to run DOCK in parallel you have to use a slightly different build of DOCK6 called dock6.mpi. | |
| + | Message passing interface ([http://www.mpi-forum.org/docs/docs.html MPI]) is basically a program that allows programs like DOCK to run in parallel. | ||
| − | + | So, make another file called qsub.vs.csh with the contents: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
#!/bin/tcsh | #!/bin/tcsh | ||
| − | + | #PBS -l nodes=4:ppn=2 | |
| − | # | + | #PBS -l walltime=24:00:00 |
| − | + | #PBS -N screen | |
| − | + | #PBS -o qsub.log | |
| − | + | #PBS -j oe | |
| − | + | #PBS -V | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | cd /nfs/user03/username/AMS536/dock-tutorial/07.virtscreen | |
| − | + | mpirun -np 8 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i dockvs.in -o dockvs.out | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | As you can see there are two major changes: | |
| − | == | + | #PBS -l nodes=4:ppn=2 #Use 4 nodes, and 2 processors per node, so 8 processors |
| − | |||
| − | + | Note: since one processor is used to distribute the processes, this will run DOCK as 7 (n-1) parallel processes. | |
| − | + | mpirun -np 8 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i dockvs.in -o dockvs.out #this line uses mpi to run dock.mpi on multiple processors | |
| − | + | And then we can run: | |
| − | + | qsub qsub.vs.csh | |
Latest revision as of 17:26, 14 January 2015
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2013, using DOCK v6.6. It is assumed that users of this tutorial have a basic understanding of the command line and are able to navigate a Unix operating system.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
Orotodine Monophosphate Decarboxylase
The protein receptor which is the subject of this tutorial is orotodine monophosphate decarboxylase (OMP), a homodimeric protein from the organism Methanobacterium thermoautotrophicum. OMP is involved in the biosynthesis of several pyrimidines including uridine monophosphate (UMP), the ligand used in this tutorial. UMP is a pyrimidine-based nucleotide monomer of RNA. The structure used for this tutorial can be found at the Protein Data Bank under accenssion number 1LOQ.
Organizing Directories
While performing docking, it is convenient to create a project directory (here we call it ~AMS536/dock-tutorial/), and adopt a standard directory structure / naming scheme, so that files are easy to find and identify. For this tutorial, we will use something similar to the following:
~username/AMS536/dock-tutorial/00.files/
/01.dockprep/
/02.surface-sphgen/
/03.box-grid/
/04.dock/
/05.mini-virtual-screen/
/06.database-filter/
/07.virtual-screen/
Before beginning the tutorial, it may be easiest to make all of the sub-directories at the same time. From here on in, these subdirectories will be referred to as ~00.files/, ~01.dockprep/ etc. In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '1LOQ'.
II. Preparing the Receptor and Ligand
Downloading the PDB file (1LOQ)
Go to PDB homepage (http://www.rcsb.org/pdb/home/home.do ) enter the protein ID (1LOQ) in the search bar, click Download Files in the top-right of the webpage, then select PDB File (text). In the new window, save the file in your ~00.files/ folder.
Preparing the ligand and receptor in Chimera
NOTE: If you are accessing files remotely (i.e. through a terminal) and want to use a visualization program such as Chimera, first transfer the files to your local computer, run Chimera, and transfer the files back.
In this section, we will create four new files and save them in the ~01.dockprep/ folder:
1LOQ.dockprep.mol2 (complete system with hydrogens and charges) 1LOQ.receptor.mol2 (the receptor alone) 1LOQ.receptor.noH.pdb (the receptor alone, without hydrogens) 1LOQ.ligand.mol2 (the ligand alone)
To prepare these files, first make a copy of the original PDB file in the ~00.files/ folder (named, for example, 1LOQ.copy.pdb) and open it with VIM ($ vim 1LOQ.copy.pdb). Because the residue name of the ligand (U) will give us some problems when assigning charges, change the residue name "U" to "LIG" starting at line 2082. Here is an example command that will change all instances of " U" into "LIG", while preserving the correct spacing. Note there are two blank spaces before U in the following expression:
%s/ U/LIG/gc
For this command, g is short for global and c is short for check with the user before making the change.
Next, open up the PDB file (1LOQ.copy.pdb) in Chimera. To delete water molecules and other ligands, click Tools -> Structure Editing -> Dock Prep. Check all boxes and click Okay to the end. Alternatively, the waters can be deleted manually by choosing Select -> Residue -> HOH, then go to Actions -> Atoms/Bonds -> Delete. Hydrogen atoms can be added manually by choosing Tools -> Structure Editing -> Add H.
Next, to add charges to the ligand and receptor, go to Select -> Residue -> LIG, then go to Tools -> Structure Editing -> Add Charge. Choose AMBER ff99SB as the charge model, click Okay, and when prompted chose AM1-BCC charges for the ligand, and make sure the Net Charge is set to -1. (You must consider the chemistry of the ligand when assigning a charge state).
Finally, save this file as 1LOQ.dockprep.mol2 into your ~01.dockprep/ folder.
Generating the final files
To create the receptor file: Open 1LOQ.dockprep.mol2, click Select -> Residue -> LIG. Then click Actions -> Atoms/Bonds -> Delete. Save the file as 1LOQ.receptor.mol2.
To create the receptor file with no hydrogen atoms: Open 1LOQ.receptor.mol2, click Select -> Chemistry -> Element -> H, then chose Actions -> Atoms/Bonds -> Delete. Save the file as 1LOQ.receptor.noH.pdb.
To create the ligand file: Open 1LOQ.dockprep.mol2, click Select -> Residue -> LIG, then click Select -> Invert, then chose Actions -> Atoms/Bonds -> Delete. Save the file as 1LOQ.ligand.mol2.
III. Generating Receptor Surface and Spheres
Receptor Surface
The next section of this tutorial will be performed in your ~02.surface-sphgen/ folder.
In order to generate the receptor surface one can perform the following steps (working in Chimera):
- Go File -> Open and choose the PDB file of the protein containing no hydrogens (1LOQ.receptor.noH.pdb) from 01.dockprep
- Further, Actions -> Surface -> Show
- Go Tools -> Structure Editing -> Write DMS in order to obtain a dms file, which we will need while placing spheres
- In the new window save the surface as 1LOQ.receptor.dms
Placing Spheres
For more information SPHGEN, please consult the DOCK online owners manual at:
http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm
In order to place the spheres on the receptor surface, the following steps are required:
1. Create a file called INSPH (an input file we will need later) and fill it out as follows (parameter descriptions included to the right), then save it.
1LOQ.receptor.dms R X 0.0 4.0 1.4 1LOQ.receptor.sph
Here is a line-by-line break down on what each line does:
- 1LOQ.receptor.dms - molecular surface file that we got from previous step
- R - sphere outside of surface (R) or inside surface (L)
- X - specifies subset of surface points to be used (X=all points)
- 0.0 - prevents generation of large spheres with close surface contacts(defalut=0.0)
- 4.0 - maximum sphere radius in angstroms (default=4.0)
- 1.4 - minimum sphere radius in angstroms (default=radius of probe)
- 1LOQ.receptor.sph - clustered spheres file that we want to generate
2. Run the sphgen program from the command line:
sphgen -i INSPH -o OUTSPH
OUTSPH is the output file containing the information on the sphgen run, while 1LOQ.receptor.sph is the file which contains the spheres.
3. (optional) If one wants to have a look at the spheres received as an outcome of sphgen, one should run showsphere, by typing in the command line:
showsphere
The following questions will be prompted on the command line:
Enter name of sphere cluster file: Enter cluster number to process (<0 = all): Generate surfaces as well as pdb files (<N>/Y)? Enter name for output file prefix: Process cluster 0 (contains ALL spheres) (<N>/Y)?
As the questions have been answered the program will generate the spheres, writing the data in a PDB file, afterwards the spheres may be observed by opening this file in Chimera.
4. Run the sphere_selector program from the command line as follows:
sphere_selector 1LOQ.receptor.sph ../01.dockprep/1LOQ.ligand.mol2 8.0
This program selects the spheres within a user-defined radius (8.0 here) of a target molecule from the previously obtained file 1LOQ.receptor.sph. As a result a new file selected_spheres.sph will be generated. Again, the spheres can and should be visualized by using showsphere and Chimera.
5. Run showsphere in order to visualize the spheres:
showsphere
When prompted on the command line, answering the questions as follows:
selected_spheres.sph -1 N output_spheres_selected N
- Launch Chimera, choose File -> Open, choose 1LOQ.receptor.noH.pdb
- Go File -> Open, choosing output_spheres_selected.pdb
- Go Select -> Residue -> SPH
- Finally, Actions -> Atoms/Bonds -> sphere
The picture like one placed below will be obtained afterwards.
IV. Generating Box and Grid
This section of the tutorial will be performed in your ~03.box-grid/ folder. For more information GRID, please consult the DOCK online owners manual at:
http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm
Box
We will create a box around the binding site to define the boundaries of ligand sampling and aid in energy calculations. As a general rule, we will create a box around the spheres (from the previous step) with margins of at least 8.0 Angstroms on all sides. You first need to create a showbox.in file to generate the box:
vim showbox.in
Edit this file as following:
Y # Yes, generate a box 8.0 # box margin ../02.surface-sphgen/selected_spheres.sph # input sphere files 1 # which sphere cluster to use 1LOQ.box.pdb # output file name
Note: Do not include the comments in the showbox.in file. To run the program showbox, give the name of the program on the command line and provide the showbox.in file:
showbox < showbox.in
Grid
Grids are used as a method to speed up evaluation of the energy scoring function by pre-computing electrostatic and VDW potential energies on the receptor at defined grid points within the box. VDW parameters are stored in a file that was installed with DOCK, and we will find it convenient to copy it and two other files to our ~00.files/ folder. In this class, DOCK was installed in the following location:
/home/wjallen/AMS536/local/dock6/
You can determine the path to your DOCK installation by typing:
which dock6
From that location, copy the following three files to your local ~00.files/ folder:
/home/wjallen/AMS536/local/dock6/parameters/vdw_AMBER_parm99.def /home/wjallen/AMS536/local/dock6/parameters/flex.defn /home/wjallen/AMS536/local/dock6/parameters/flex_drive.tbl
Use grid program to generate the grid and answer the questions as following:
touch grid.in grid -i grid.in
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../01.dockprep/1LOQ.receptor.mol2 box_file ../03.box-grid/1LOQ.box.pdb vdw_definition_file ../00.files/vdw_AMBER_parm99.defn score_grid_prefix 1LOQ.grid
V. Docking a Single Molecule for Pose Reproduction
This part of the tutorial will be performed in your ~04.dock/ folder. For more information DOCK, please consult the DOCK online owners manual at:
http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm
Rigid Docking
When docking a molecule rigidly in DOCK, no conformations or rotamers will be sampled other than the input configuration which, in our case, was derived from the crystal structure. This will be much faster that flexible docking, but it is not appropriate for virtual screening because the docked conformation of compounds from a library is generally not known. First create an empty input file for docking:
touch dock.in
Then execute DOCK6:
dock6 -i dock.in
You will be prompted for answers to many questions. For the rigid docking experiments, suggested parameters are as follows:
ligand_atom_file ../01.dockprep/1LOQ.ligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphgen/selected_spheres.sph use_internal_energy no flexible_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1LOQ.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix output write_orientations no num_scored_conformers 1 rank_ligands no
With these input parameters, the rigid docking experiment will result in a single scored ligand pose. Open Chimera to visualize the docking result. After opening the receptor file in surface view, open the ligand file (named "output_scored.mol2") by going under Tools -> Surface/Binding Analysis -> ViewDock.
Flexible Docking
Flexible docking will take longer, but is necessary for virtual screening. For flexible docking, several input parameters need to be changed, and these changes will generate additional questions prompted to the user. Some changes to parameters are shown below:
calculate_rmsd yes flexible_ligand yes use_internal_energy yes minimize_ligand yes num_scored_conformers 10
Once these changes have been made to the input file, execute DOCK6 again:
dock6 -i dock.in
This flexible ligand protocol can be used with multiple ligand input files. The docked ligand poses can be compared using RMSD (Root Mean Squared Deviation). RMSD is a measurement of the distance between atoms on the original crystal structure ligand with the newly generated ligand conformation. An RMSD under 2 Angstroms would indicate a good conformation. Below are two examples of a final docked ligand in the receptor binding site pocket. The worst and best energy scored ligands are shown below. The original ligand is in cyan and the docked ligand is colored in heteroatom.
VI. Virtual Screening
Virtual Screening Introduction
A virtual screen of various ligand allows for the comparison of both qualitative (e.g. position in binding site) and quantitative (e.g. energy scores) data pertaining to the each screened ligand with an originally docked molecule. Virtual screening is often used as a method to cut the cost of experimentation by narrowing down the ligands within a database and predicting which will exhibit the most favorable binding to a specific protein (with a pre-determined .grid file).
Running the Database Filter
When first beginning the virtual screen, a filter should be specified in order save computational power by only screening potentially effective ligands within the directory ~06.database-filter/. This involves limiting several variables of ligands specified in a .mol2 file. To limit the ligands from the original database, a file "dock_filter.in" is created and executed using DOCK. Note that in this example the file "cdiv_p0.0.mol2" is a multi mol2 file containing thousands of ligands from the CHEMDIV vendor which was previously downloaded from the UCSF ZINC database (https://zinc.docking.org/about/ucsf). ZINC is a great resource of purchasable compounds for virtual screening. Here are the contents of the file:
ligand_atom_file /home/wjallen/AMS536/multi-mol2-files/cdiv_p0.0.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter yes dbfilter_max_heavy_atoms 50 dbfilter_min_heavy_atoms 0 dbfilter_max_rot_bonds 10 dbfilter_min_rot_bonds 0 dbfilter_max_molwt 9999.0 dbfilter_min_molwt 0.0 dbfilter_max_formal_charge -1.0 dbfilter_min_formal_charge -2.0 orient_ligand no use_internal_energy no flexible_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix filtered write_orientations no num_scored_conformers 1 rank_ligands no
In this file some important characteristics of the ligands that are limited are dbfilter_max_heavy_atoms, dbfilter_max_rot_bonds, dbfilter_max_molwt, and both dbfilter_max_formal_charge and dbfilter_min_formal_charge. As show in the text of the file, a file called filtered_scored.mol2 will be created with all of the ligands from the original database that fit into the specified limits of number of heavy atoms, number of rotatable bonds, molecular weight, and formal charge. Note: The max and min formal charge specified in this file (-1 to -2) are an extremely limited range, and are only used in this tutorial as an illustration.
Running the Virtual Screen
After creating a new directory, ~07.virtual-screen/, a dock.in file must be created. So as not to confuse the virtual screen file with the original dock.in of the docked ligand, the new file created is called dock_vs.in. Running this file in dock6 allows us to fill in the important details of the file that terminate in a virtual screening of the input ligand.
ligand_atom_file ../06.database-filter/filtered_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphgen/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no use_internal_energy yes internal_energy_rep_exp 12 flexible_ligand yes user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 use_clash_overlap no write_growth_tree no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1LOQ.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix output write_orientations no num_scored_conformers 1 rank_ligands no
A few of the questions addressed in the terminal window while running a virtual screen are as follows:
ligand_atom_file - here we have specified the new file created after filtering the initial ligand database file
calculate_rmsd - we select the default <no> because since we are screening a library of compounds, there will be no reference position to compare them to
orient_ligand - here we would like to the have the ligand oriented in the binding pocket
receptor_site_file - this is our selected_spheres.sph file from earlier
internal_energy_rep_exp - here we select 12 rather than 9 because using a repulsive exponent of 12 gives us a slightly steeper rising potential as radius between the atoms falls below the optimal distance (sigma)
flexible_ligand - when performing a virtual screen of a random database it is always advisable to set the ligands to flexible
min_anchor_size - ensures that major features on the molecules are used as anchors and not just simple small groups
score_molecules - we always want to score the molecules of a virtual screen in order to determine which are the best fit for the binding pocket
grid_score_primary - we want the grid score to be the primary method of scoring the screened ligands
grid_score_grid_prefix - the input for the .grid file created earlier
atom_model - we select the all atom model
num_scored_conformers - we only want to produce one scored conformer from each screened ligand because we only want the best conformations
Note that if you are performing a small sample virtual screening it is possible to run it on a local computer in a reasonable amount of time; however, if you are running the virtual screening to a very large scale (i.e. hundreds or thousands of ligands) you will need to use several nodes in Seawulf to produce results in a timely fashion.
Analyzing the Results
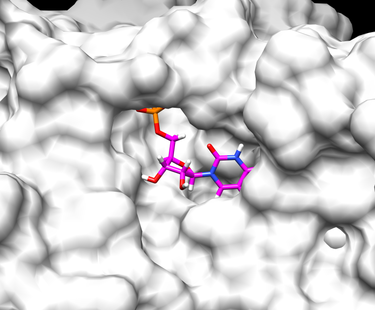
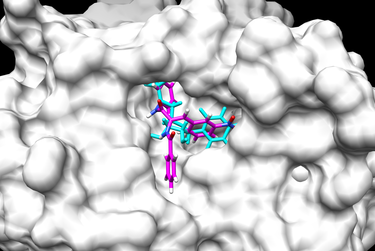
When your virtual screening has run completely and you are ready to analyze the results, open Chimera. In Chimera you can prepare your protein by opening 1LOQ.receptor.noH.pdb and choosing Actions -> Surface -> show. You may also wish to open up the original ligand file 1LOQ.ligand.mol2 in order to show the position of the original ligand/substrate in the receptor's active site.
To view the results of your virtual screening go to Tools -> Surface/Binding Analysis -> ViewDock and from their you can select your virtual screen output file. In the "ViewDock" pop-up window you can then select Column --> Show --> Grid Score which will add a column to the ViewDock window with the grid score of each ligand. You can double-click on the column heading Grid Score to sort the ligands based on their grid scores.
[Chimera image1] [Chimera image2]
VII. Running DOCK in Serial and in Parallel on Seawulf
The Seawulf Cluster is a 470-processor Linux Cluster capable of highly parallel processing. This parallel processing allows dock virtual screens to be completed in a fraction of the time as a single processor.
If you are docking multiple ligands, you can use more than one processor in parallel mode, but you should never use more processors than you have ligands. Before we can run DOCK on Seawulf, we need to copy the proper files from Herbie to Seawulf. If we CD into the AMS536 folder we can use the following command from the mathlab computer to copy all of the dock-tutorial files
scp -r /dock-tutorial/ username@herbie.mathlab.sunysb.edu:~/AMS536/
Now we have all of our DOCK preparation files and folders on the seawulf cluster.
Running DOCK in Serial on a Single Processor
Running on a single processor is very similar to running dock on the mathlab computer.
If you make a file called qsub.csh with the text:
#!/bin/tcsh #PBS -l nodes=1:ppn=1 #PBS -l walltime=01:00:00 #PBS -N dock6 #PBS -M user@ic.sunysb.edu #PBS -j oe #PBS -o pbs.out cd /nfs/user03/zfoda/AMS536/dock-tutorial/07.virtscreen /nfs/user03/wjallen/local/dock6/bin/dock6 -i dock.in -o dock.out
An explanation of the commands:
#!/bin/tcsh #Execute script with tcsh #PBS -l nodes=1:ppn=1 #Use one node, and one processor per node, so one single processor #PBS -l walltime=01:00:00 #Allow 1 hour for your job run #PBS -N dock6 #Name of your job #PBS -M user@ic.sunysb.edu #Get an email notifying you when your job is completed #PBS -j oe #Combine the output and error streams into a single output file #PBS -o pbs.out #Name of your output file cd /nfs/user03/zfoda/AMS536/dock-tutorial/07.virtscreen #Change to your home directory and folder with dock files /nfs/user03/wjallen/local/dock6/bin/dock6 -i dock.in -o dock.out #Specifies path to dock executable and provide input and output filenames
To submit the experiment use the command:
qsub qsub.csh
You will have submitted a DOCK experiment to the seawulf queue.
See also PBS commands.
Running DOCK in Parallel using MPI
In order to run DOCK in parallel you have to use a slightly different build of DOCK6 called dock6.mpi. Message passing interface (MPI) is basically a program that allows programs like DOCK to run in parallel.
So, make another file called qsub.vs.csh with the contents:
#!/bin/tcsh #PBS -l nodes=4:ppn=2 #PBS -l walltime=24:00:00 #PBS -N screen #PBS -o qsub.log #PBS -j oe #PBS -V cd /nfs/user03/username/AMS536/dock-tutorial/07.virtscreen mpirun -np 8 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i dockvs.in -o dockvs.out
As you can see there are two major changes:
#PBS -l nodes=4:ppn=2 #Use 4 nodes, and 2 processors per node, so 8 processors
Note: since one processor is used to distribute the processes, this will run DOCK as 7 (n-1) parallel processes.
mpirun -np 8 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i dockvs.in -o dockvs.out #this line uses mpi to run dock.mpi on multiple processors
And then we can run:
qsub qsub.vs.csh