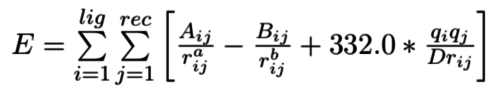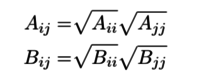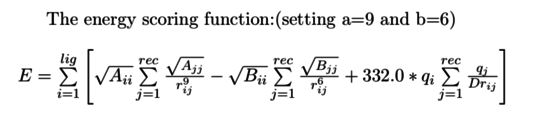Difference between revisions of "Energy Scoring Method in Grid"
Stonybrook (talk | contribs) (Created page with "This is a test page") |
Stonybrook (talk | contribs) |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | The general form of energy scoring function: | |
| + | [[Image:1.png|thumb|center|500px|Energy scoring function]] | ||
| + | |||
| + | It's easy to see that the Coulumb potential Energy term could be separated into receptor and ligand part. If the Lennard-Jones one could be separated, it will be easy to precalculate the potential of each grid point in docking region and speed up the docking process. | ||
| + | |||
| + | Computational chemist took the assumption that the coefficients for Lennard-Jones part in energy scoring function are also separable. | ||
| + | [[Image:2.png|thumb|center|200px|Lennard-Jones coefficients]] | ||
| + | |||
| + | The new form of energy scoring function like this: | ||
| + | |||
| + | [[Image:3.png|thumb|center|550px|Energy score function]] | ||
| + | |||
| + | |||
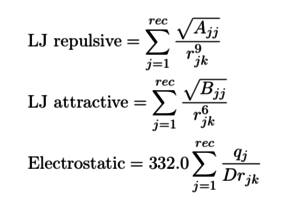
| + | So each time, when you run the grid calculation, you get the grid value in each grid point | ||
| + | |||
| + | [[Image:4.png|thumb|center|300px|potential energy at each grid point]] | ||
| + | |||
| + | and save them in your *.nrg file. | ||
| + | |||
| + | In dock process, the program generate the energy of certain ligand atom by statistically summarizing the closest eight gird points' value. The energy score for a ligand will be the total of all its atoms | ||
Latest revision as of 14:24, 5 March 2015
The general form of energy scoring function:
It's easy to see that the Coulumb potential Energy term could be separated into receptor and ligand part. If the Lennard-Jones one could be separated, it will be easy to precalculate the potential of each grid point in docking region and speed up the docking process.
Computational chemist took the assumption that the coefficients for Lennard-Jones part in energy scoring function are also separable.
The new form of energy scoring function like this:
So each time, when you run the grid calculation, you get the grid value in each grid point
and save them in your *.nrg file.
In dock process, the program generate the energy of certain ligand atom by statistically summarizing the closest eight gird points' value. The energy score for a ligand will be the total of all its atoms