Difference between revisions of "2017 DOCK tutorial 1 with PDB 4QMZ NEW"
Stonybrook (talk | contribs) (→III. Generating Receptor Surface and Spheres) |
Stonybrook (talk | contribs) (→Rescoring Docked Molecules) |
||
| (25 intermediate revisions by the same user not shown) | |||
| Line 152: | Line 152: | ||
| − | Then we need to select the spheres pertinent to our docking experiment. Usually these | + | Then we need to select the spheres pertinent to our docking experiment. Usually these spheres will be the closest N spheres to the native ligand molecule. |
Run | Run | ||
sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 10.0 | sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 10.0 | ||
| − | This command will select all of the spheres within | + | This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph |
To visualize the spheres using Chimera as previously done: | To visualize the spheres using Chimera as previously done: | ||
| Line 182: | Line 182: | ||
This input designates to dock that: | This input designates to dock that: | ||
| − | we want to create a box | + | we want to create a box, |
| − | the box length should be 8.0 Angstroms | + | the box length should be 8.0 Angstroms, |
| − | use the selected spheres in the file | + | use the selected spheres in the file designated, |
| − | output the box to the file specified | + | output the box to the file specified |
| Line 220: | Line 220: | ||
this script should output verbosely to the terminal and produce 2 output files, grid.bmp and grid.nrg, both binary files. | this script should output verbosely to the terminal and produce 2 output files, grid.bmp and grid.nrg, both binary files. | ||
| + | |||
| + | grid -i grid.in > gridinfo.out | ||
| + | |||
| + | Check the standard output file gridinfo.out to ensure that individual charges on charged residues are integer values (-1 or +1). | ||
| + | |||
| + | University of California at San Francisco, DOCK 4.0.1 | ||
| + | |||
| + | __________________Job_Information_________________ | ||
| + | launch_time Wed Feb 8 17:05:50 2017 | ||
| + | host_name unknown | ||
| + | memory_limit -1 | ||
| + | working_directory /home/campus.stonybrook.edu/ronassar/MySBFiles/projects/dock_tutorial/03.box-grid2_savegridoutput | ||
| + | user_name ronassar | ||
| + | |||
| + | ________________General_Parameters________________ | ||
| + | compute_grids yes | ||
| + | grid_spacing 0.4 | ||
| + | output_molecule no | ||
| + | |||
| + | ________________Scoring_Parameters________________ | ||
| + | contact_score no | ||
| + | energy_score yes | ||
| + | energy_cutoff_distance 9999 | ||
| + | atom_model a | ||
| + | attractive_exponent 6 | ||
| + | repulsive_exponent 12 | ||
| + | distance_dielectric yes | ||
| + | dielectric_factor 4 | ||
| + | bump_filter yes | ||
| + | bump_overlap 0.75 | ||
| + | |||
| + | ____________________File_Input____________________ | ||
| + | receptor_file ../01.dockprep/4qmz.rec.mol2 | ||
| + | box_file ../03.box-grid/4qmz.box.pdb | ||
| + | vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | |||
| + | ____________________File_Output___________________ | ||
| + | score_grid_prefix grid | ||
| + | |||
| + | |||
| + | |||
| + | Reading in coordinates of receptor. | ||
| + | CHARGED RESIDUE MET : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE ASP : -1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE ARG : 1.000 | ||
| + | CHARGED RESIDUE LYS : 1.000 | ||
| + | CHARGED RESIDUE GLU : -1.000 | ||
| + | CHARGED RESIDUE SER : -1.000 | ||
| + | |||
| + | Total charge on 4qmz_rec_leap.pdb : -5.000 | ||
| + | |||
| + | Reading in grid box information. | ||
| + | Box center of mass : -1.912 0.054 14.111 | ||
| + | Box dimensions : 39.856 41.687 38.838 | ||
| + | Number of grid points per side [x y z] : 101 106 99 | ||
| + | Total number of grid points : 1059894 | ||
| + | |||
| + | Generating scoring grids. | ||
| + | Percent of protein atoms processed : 0 | ||
| + | Percent of protein atoms processed : 10 | ||
| + | Percent of protein atoms processed : 20 | ||
| + | Percent of protein atoms processed : 30 | ||
| + | Percent of protein atoms processed : 40 | ||
| + | Percent of protein atoms processed : 50 | ||
| + | Percent of protein atoms processed : 60 | ||
| + | Percent of protein atoms processed : 70 | ||
| + | Percent of protein atoms processed : 80 | ||
| + | Percent of protein atoms processed : 90 | ||
| + | Percent of protein atoms processed : 100 | ||
| + | Writing general grid info to grid.bmp | ||
| + | Writing bump grid to grid.bmp | ||
| + | Writing energy grids to grid.nrg | ||
| + | Writing attractive VDW energy grid | ||
| + | Writing repulsive VDW energy grid | ||
| + | Writing electrostatic energy grid | ||
| + | |||
| + | Finished calculation. | ||
==V. Docking a Single Molecule for Pose Reproduction== | ==V. Docking a Single Molecule for Pose Reproduction== | ||
| Line 225: | Line 381: | ||
Create or enter directory 4, 04.dock | Create or enter directory 4, 04.dock | ||
| − | + | ===Minimization=== | |
create min.in | create min.in | ||
| Line 294: | Line 450: | ||
This structure can be visualized using chimera and loading the surface into chimera along with the minimized and unminimized ligand structures: | This structure can be visualized using chimera and loading the surface into chimera along with the minimized and unminimized ligand structures: | ||
| + | [[Image:image_min.png|thumb|center|1000px|4qmz with ligand before and after minimization]] | ||
| + | The RMSD of the minimized ligand to crystal structure ligand is 0.297 A. | ||
| − | At this point we are going to calculate the van der | + | At this point we are going to calculate the van der waals and electrostatic footprints for the unminimized and minimized ligand structures in relation to the receptor active site. |
create the input file: footprint.in | create the input file: footprint.in | ||
| Line 370: | Line 528: | ||
The output will look something like this: | The output will look something like this: | ||
| + | |||
| + | [[Image:figure_fps_minimized.png|thumb|center|1000px|Footprint comparison between unminimized and minimized ligand structure]] | ||
| + | |||
| + | Notice the vdW clashes are now reduced (vdW positive peaks disappeared). | ||
| + | |||
| + | |||
| + | ===Rigid Docking=== | ||
| + | |||
| + | To help identify any errors in preparing the ligand and receptor files, a rigid docking, fixed growth docking and flexible docking runs will be performed. This is a check that the program can put the ligand into the crystal structure pose before proceeding. | ||
| + | |||
| + | Create an input file 'rigid.in' for the rigid docking run with the following parameters: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../01.dockprep/4qmz.lig.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../02.surface-spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../03.box-grid/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /opt/AMS536/dock6/parameters/flex.defn | ||
| + | flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4qmz.rigid_norestraint | ||
| + | write_orientations no | ||
| + | num_scored_conformers 50 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | Outputs the file "4qmz.rigid_norestraint" the no restraint refers to the option of the parameter simplex_restraint_min in the input file. This removes the tether when doing the minimization. The best scoring structure had an RMSD of 1.??? to the native ligand pose. | ||
| + | |||
| + | [[Image:rigiddockingresults.png|thumb|center|800px|Rigid docking results]] | ||
| + | |||
| + | ===Fixed Anchor Docking=== | ||
| + | |||
| + | In fixed anchor docking, the ligand location is fixed in the active site and the rotatable bonds space is searched (i.e angles are flexible). | ||
| + | |||
| + | Create an input file 'fad.in' for the rigid docking run with the following parameters: | ||
| + | Notice the search algorithm is now rigidset to flex (for flexible) and orient option is set to no (for fixing the anchor in place). | ||
| + | |||
| + | conformer_search_type flex | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../01.dockprep/4qmz.lig.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../02.surface-spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../03.box-grid/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /opt/AMS536/dock6/parameters/flex.defn | ||
| + | flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4qmz.fad | ||
| + | write_orientations no | ||
| + | num_scored_conformers 100 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | [[Image:fixedanchorresults.png|thumb|center|800px|Fixed anchor docking results]] | ||
| + | |||
| + | ===Flexible Docking=== | ||
| + | |||
| + | In flexible docking, the ligand location and rotatable bonds are search. | ||
| + | |||
| + | Create an input file 'flex.in' for the flexible docking run with the following parameters: | ||
| + | Notice the search algorithm is now set to flex (for flexible) and orient option is set to yes (for fixing the anchor in place). | ||
| + | |||
| + | conformer_search_type flex | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../01.dockprep/4qmz.lig.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../02.surface-spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../03.box-grid/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /opt/AMS536/dock6/parameters/flex.defn | ||
| + | flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4qmz.flex | ||
| + | write_orientations no | ||
| + | num_scored_conformers 100 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | [[Image:flexibledockingresults.png|thumb|center|800px|Flexible docking results]] | ||
==VI. Virtual Screening== | ==VI. Virtual Screening== | ||
| + | |||
| + | At this point we will begin docking potential inhibitors into the active site of 4qmz. The compound library we will use contains 25,000 molecules and was previously prepared for another docking experiment. The compound library contains molecules with rotatable bonds number between 7 and 15. The library used is titled small_ligand_library.mol2, this library can be found at /gpfs/projects/AMS536/virtual_screen_library on seawulf. | ||
| + | |||
| + | We began by copying all of our local directories outlined in the first section to a project directory on seawulf to allow use to expedite the docking process over the 25,000 compounds in our test library. | ||
| + | |||
| + | We will create a virtual-screen.in file that looks like the following: | ||
| + | |||
| + | conformer_search_type flex | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file small_ligand_library.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../02.surface-spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../03.box-grid/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4qmz.virtualscreen | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | This script is submitted to the seawulf cluster using a script called screen.sh which contains the following: | ||
| + | |||
| + | #! /bin/tcsh | ||
| + | #PBS -l nodes=4:ppn=28 | ||
| + | #PBS -l walltime=24:00:00 | ||
| + | #PBS -o jobstatus.out | ||
| + | #PBS -e joberror.err | ||
| + | #PBS -N large_vs | ||
| + | #PBS -V | ||
| + | #PBS -q long | ||
| + | cd $PBS_O_WORKDIR | ||
| + | mpirun -np 112 dock6.mpi -i virtual-screen.in -o virtualscreen1.out | ||
| + | |||
| + | |||
| + | This screen.sh input specifies that we will use 112 processors to dock our virtual library. We have a wall clock for the job of 24 hours and we will output jobstatus.out, joberror.err and virtualscreen1.out. In addition to these output files there is an output for the work performed by each processor individually. | ||
| + | |||
| + | ===Cartesian Minimization of the docked molecules=== | ||
| + | |||
| + | Now we will do cartesian minimization of the docked molecules. Cartesian here is referring to the receptor being explicitly present during the minimization. | ||
| + | |||
| + | Write the minimization input script minimize-vs.in: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file 4qmz.virtualscreen_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary yes | ||
| + | continuous_score_secondary no | ||
| + | cont_score_rec_filename ../01.dockprep/4qmz.rec.mol2 | ||
| + | cont_score_att_exp 6 | ||
| + | cont_score_rep_exp 12 | ||
| + | cont_score_rep_rad_scale 1 | ||
| + | cont_score_use_dist_dep_dielectric yes | ||
| + | cont_score_dielectric 4.0 | ||
| + | cont_score_vdw_scale 1 | ||
| + | cont_score_es_scale 1 | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4qmz.virtualscreen1.minimized | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Submit the minimization job using with the script "job-min.sh" following lines: | ||
| + | |||
| + | #!/bin/tcsh | ||
| + | #PBS -l nodes=16:ppn=28 | ||
| + | #PBS -l walltime=12:00:00 | ||
| + | #PBS -o jobstatusmin.out | ||
| + | #PBS -e jobstatusmin.err | ||
| + | #PBS -N chunk0min | ||
| + | #PBS -V | ||
| + | #PBS -q medium | ||
| + | module load intel/mpi/64/2017/0.098 | ||
| + | cd $PBS_O_WORKDIR | ||
| + | mpirun -np 448 /gpfs/projects/AMS536/zzz.programs/dock6/bin/dock6.mpi -i minimize-vs.in -o minimize-vs.out | ||
| + | |||
| + | |||
| + | At this point it would be good to rank our docked ligands by score and extract the 100 best scoring (100 lowest scoring, most negative) ligands to investigate further. | ||
| + | |||
| + | |||
| + | ===Rescoring Docked Molecules=== | ||
| + | |||
| + | Dock has several built in scoring functions. In an effort to utilize these alternative scoring functions it is worthwhile to rescore the results from the initial virtual screen that were collected. The virtual screen will be rescored using footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score. Each of these scoring functions focuses in on a different chemical aspect of the ligand in question. The rescore.in script for this run can be found here: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../05.virtual- screen/4qmz.virtualscreen1.minimized_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score no | ||
| + | descriptor_use_multigrid_score no | ||
| + | descriptor_use_continuous_energy no | ||
| + | descriptor_use_footprint_similarity yes | ||
| + | descriptor_use_pharmacophore_score yes | ||
| + | descriptor_use_tanimoto yes | ||
| + | descriptor_use_hungarian yes | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_fps_use_footprint_reference_mol2 yes | ||
| + | descriptor_fps_footprint_reference_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 | ||
| + | descriptor_fps_foot_compare_type Euclidean | ||
| + | descriptor_fps_normalize_foot no | ||
| + | descriptor_fps_foot_comp_all_residue yes | ||
| + | descriptor_fps_receptor_filename ../01.dockprep/4qmz.rec.mol2 | ||
| + | descriptor_fps_vdw_att_exp 6 | ||
| + | descriptor_fps_vdw_rep_exp 12 | ||
| + | descriptor_fps_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_dielectric 4.0 | ||
| + | descriptor_fps_vdw_fp_scale 1 | ||
| + | descriptor_fps_es_fp_scale 1 | ||
| + | descriptor_fps_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename ../04.dock/4qmz.lig.min_scored.mol2 | ||
| + | descriptor_hungarian_ref_filename ../04.dock/4qmz.lig.min_scored.mol2 | ||
| + | descriptor_hungarian_matching_coeff -5 | ||
| + | descriptor_hungarian_rmsd_coeff 1 | ||
| + | descriptor_volume_reference_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 | ||
| + | descriptor_volume_overlap_compute_method analytical | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
| + | descriptor_weight_fingerprint_tanimoto -1 | ||
| + | descriptor_weight_hungarian 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn | ||
| + | pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | |||
| + | From here the results should be sorted according the score based on the scoring function in question. A script for this can be written fairly quickly in whatever language you are most comfortable with. If you are not very experienced with computer programming, a python script developed in house can be found here that will search through the output .mol2 file and select the top N scoring molecules. These top scoring molecule s are taken from the virtual screen "top'N'_molid.txt" and finds and their coordinates saved to an output file "top'N'_molid.mol2". | ||
| + | |||
| + | rank_scores_get_coords.py: | ||
| + | |||
| + | #!/usr/bin/python | ||
| + | import sys | ||
| + | from operator import itemgetter, attrgetter, methodcaller | ||
| + | # input mol2 file as command line argument | ||
| + | f0=open(sys.argv[1],'r') ; data0 = f0.readlines() ; f0.close() #open mol2 data file with scored molecules | ||
| + | # define variables | ||
| + | nummol = 50 # number of molecules to keep after ranking by score | ||
| + | #scoretype="Grid_Score:" # scoring function to rank by | ||
| + | scoretype="Footprint_Similarity_Score:" | ||
| + | #-------------------------------------------------------------------------------------------------------------------------- | ||
| + | list_molid=[] #create a list to store molecule ids | ||
| + | list_molscores=[] #create list to store scores for each respective molecule | ||
| + | for line in data0: #reads through the input file | ||
| + | if line != "\n": #if the line is not empty continue | ||
| + | if line.split()[0]=="##########": #if the line starts with ############ then continue | ||
| + | if line.split()[1]=="Name:": #if the line has the next entry as Name: continue | ||
| + | molid = line.split()[2] #then add the molID to list_molid | ||
| + | list_molid.append(molid) | ||
| + | elif line.split()[1]==scoretype: | ||
| + | molscore = line.split()[2] | ||
| + | list_molscores.append(molscore) | ||
| + | #print list_molid | ||
| + | #print list_molscores | ||
| + | both = zip(list_molid,list_molscores) #concatenates list_molid and list_molscores into one array | ||
| + | sorted_both=sorted(both, key=itemgetter(1), reverse=True) #sorts the "both" list | ||
| + | sorted_top=[] #names a new list sorted_top5 | ||
| + | #for element in sorted_both: #print out first n entries in sorted_both | ||
| + | # print element[0], element[1] | ||
| + | o=open("top"+str(nummol)+"_molid.txt","w") #opens output file topx_molid.txt | ||
| + | for element in sorted_both[:nummol]: | ||
| + | sorted_top.append(element[0]) | ||
| + | o.write(element[0]+"\n") #stores the top nummol entries in sorted_top | ||
| + | o.close() | ||
| + | #print sorted_top | ||
| + | ## | ||
| + | ##-------------------------------------------------------------------------------------------------------------------------- | ||
| + | ## | ||
| + | ## Get coordinates of molecules of interest from scored mol2 | ||
| + | f1=open("top"+str(nummol)+"_molid.txt","r") ; data1=f1.readlines(); f1.close() # takes in sorted list of top molecules | ||
| + | f2=open(sys.argv[1],"r") ; data2=f2.readlines(); f2.close() # takes in mol2 results | ||
| + | identifier=0 # this will switch between 0 and 1. When it is 0 no line printing, when it is 1 we print the line. | ||
| + | molid_top=[] # list of molecule ids that are of interest | ||
| + | # go through the first file (has top n molecules) and append those ids to molecule ids list | ||
| + | for line in data1: | ||
| + | molid=line.split()[0] | ||
| + | molid_top.append(molid) | ||
| + | output=open("top"+str(nummol)+"_molid.mol2","w") | ||
| + | # go through the second file (the rescored mol2 file), the next commands print out the coordinates for the molecules of interest | ||
| + | for line in data2: | ||
| + | # if line != "\n": # skipping empty lines | ||
| + | if identifier==1: | ||
| + | try: | ||
| + | if line.split()[0]=="##########" and line.split()[1]=="Name:" and line.split()[2] not in molid_top: # changing the identifier when hitting a new molecule that is not of interest | ||
| + | identifier=0 | ||
| + | output.write('\n') | ||
| + | else: | ||
| + | output.write(line) # else keep the identifier at 1 and keep printing | ||
| + | except IndexError: | ||
| + | continue | ||
| + | try: | ||
| + | if identifier==0 and line.split()[0]=="##########" and line.split()[1]=="Name:" and line.split()[2] in molid_top: # change identifier to 1 when we hit molecule of interest | ||
| + | identifier=1 | ||
| + | output.write(line) | ||
| + | except IndexError: | ||
| + | continue | ||
| + | output.close() | ||
==VIII. Frequently Encountered Problems== | ==VIII. Frequently Encountered Problems== | ||
| + | |||
| + | Spending the time to ensure your starting structure is reasonable is of the utmost importance and will save you much headache moving forward in your study. Reading through the PDB file as downloaded from the protein data bank will give the researcher a lot of information regarding issues or grey area encountered by the researchers responsible for publishing the structure initially. Threading a structure through some quality control software such as tleap, chimera or another comparable software package will ensure charges are reasonable, hydrogens are appropriately placed, and there are no missing residues in the peptide chain. | ||
| + | |||
| + | A basic understanding of your enzymes active site will also be beneficial while approaching an enzymatic system from a docking perspective. Some questions to ask oneself are: does my protein have an ion (Mg, Ca, etc.) in the active site? Does my ligand make important interactions with conserved water residues? If so these waters could be kept in with the receptor to aid in discovery. The alternative would be to design an inhibitor that displaces the water molecules and supplements favorable interactions mediated by the water between the ligand and protein. | ||
Latest revision as of 18:10, 8 March 2017
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by a subsection of the AMS 536 class of 2017, using DOCK v6.8.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
4QMZ
In this tutorial we will use PDB code 4QMZ, the deposited crystal structure of MST3 in complex with SUNITINIB.
Organizing Directories
While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify.For this tutorial, we will use something similar to the following:
~username/AMS536-Spring2016/dock-tutorial/00.files/
/01.dockprep/
/02.surface-spheres/
/03.box-grid/
/04.dock/
/05.large-virtual-screen/
/06.virtual-screen/
/07.footprint/
/08.print_fps
|
In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '4QMZ'. The following sections in this tutorial will adhere to this directory structure/naming scheme.
II. Preparing the Receptor and Ligand
Download the PDB file (4QMZ)
4QMZ was moved into 00.files
4qmz.pdb was copied to raw_4qmz.pdb
raw_4qmz.pdb was opened with VI terminal editor
The header information, connect records, ions (atoms 2333 and 2334) and waters were deleted
Res 178 = TPO, or phosphonothreonine
Res 178 (TPO) was renamed to THR (Threonine) and HETATM renamed to ATOM, in addition the acanonical atoms were removed from the pdb leaving a deprotonated threonine (Atoms 1311-1314 in 4qmz.pdb)
Res B49 was renamed to LIG and made Chain B
raw_4qmz.pdb was copied twice to 4qmz_rec.pdb and 4qmz_lig.pdb
4qmz_rec.pdb was opened with VI terminal editor
LIG atoms, or chain B, was deleted and the file saved
4qmz_lig.pdb was opened with VI terminal editor
Protein atoms, or chain A, was deleted and the file saved
4qmz_rec.pdb was loaded into tleap as a quality control measure
tleap
source leaprc.protein.ff14SB
lin = loadpdb /path/to/4qmz_rec.pdb
2340 Hydrogens added, 1 heavy atom added (CSER RES 299, Chain A, OXT 12)
check lin
saveamberparm lin /path/to/4qmz_rec_leap.parm7 /path/to/4qmz_rec_leap.crd
Running the receptor through leap ensures a reasonable starting structure and can help identify obvious issues sooner rather than later.
At this point the .parm7 and .crd have been created via tleap ambpdb can be used to obtain the clean pdb 4qmz_rec_leap.pdb
ambpdb -p 4qmz_rec_leap.parm7 -c 4qmz_rec_leap.crd > 4qmz_rec_leap.pdb
Now add partial charges to the receptor and save file in .mol2 format:
open chimera
load 4qmz_rec_leap.pdb
Tools --> Structure editing --> Add charge --> AMBER ff99SB with AM1-BCC charges
File --> Save Mol2... --> 4qmz.rec.mol2
A no-hydrogen receptor pdb file will now be created:
Chimera, load 4qmz.rec.mol2
Select --> Chemistry --> element --> H
Actions --> Atoms --> Delete
File --> Save PDB... --> 4qmz.rec.noH.pdb
Ligand (4qmz_lig.pdb) will now be charged and saved in mol2 format
open chimera
load 4qmz_lig.pdb
Tools --> structure editing --> AddH
Tools --> Structure editing --> Add charge --> AMBER ff99SB with AM1-BCC charges
File --> Save mol2 --> 4qmz.lig.mol2
Placement of partial charges can be verified by examining the saved files 4qmz.lig.mol2 and 4qmz.rec.mol2.
III. Generating Receptor Surface and Spheres
Open 4qmz.rec.noH.pdb in Chimera
To generate the molecular surface:
Action --> Surface --> Show
Save the .dms file
Tools --> Structure editing --> write DMS
.dms save to 4qmz.rec.noH.dms
Create surface spheres
create input file INSPH
4qmz.rec.noH.dms
R
X
0.0
4.0
1.4
4qmz.rec.sph
line 1 designates input file line 2 designates the generated spheres will be outside the receptor surface line 3 designates that all points on the receptor will be used line 4 designates the maximum surface radius of the spheres line 5 designates the minimum surface radius of the spheres line 6 designates the output file name
run sphgen
sphgen -i INSPH -o OUTSPH
sphgen is the sphere generation program from dock -i desginates the input file: INSPH -o designates the output file
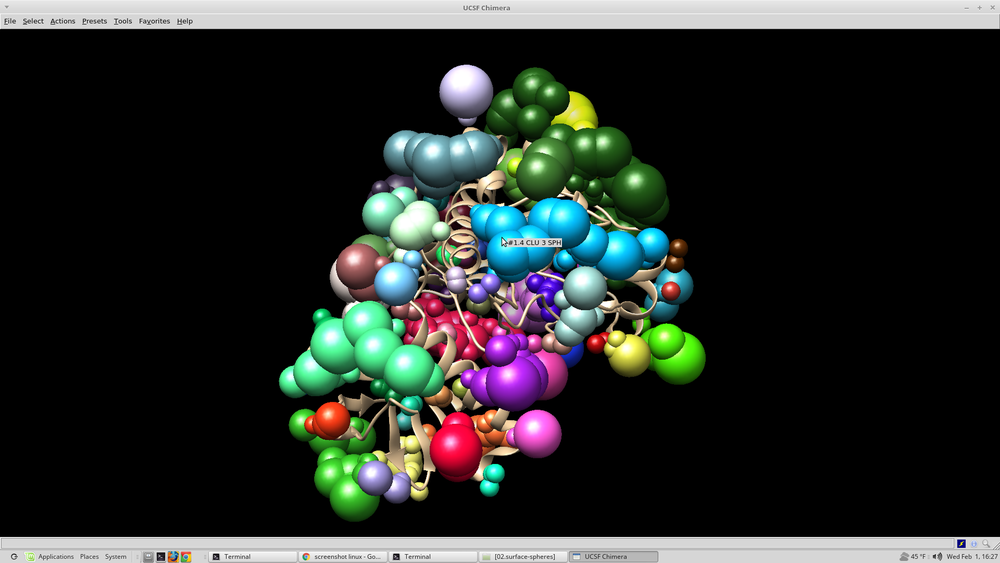
At this point it is beneficial to visualize the spheres that were created. This can be done with chimera: Open chimera from the terminal choose file --> open 4qmz.rec.mol2 choose file --> open 4qmz.rec.sph
The image that appears should resemble this:
Then we need to select the spheres pertinent to our docking experiment. Usually these spheres will be the closest N spheres to the native ligand molecule.
Run
sphere_selector 4qmz.rec.sph ../01.dockprep/4qmz.lig.mol2 10.0
This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph
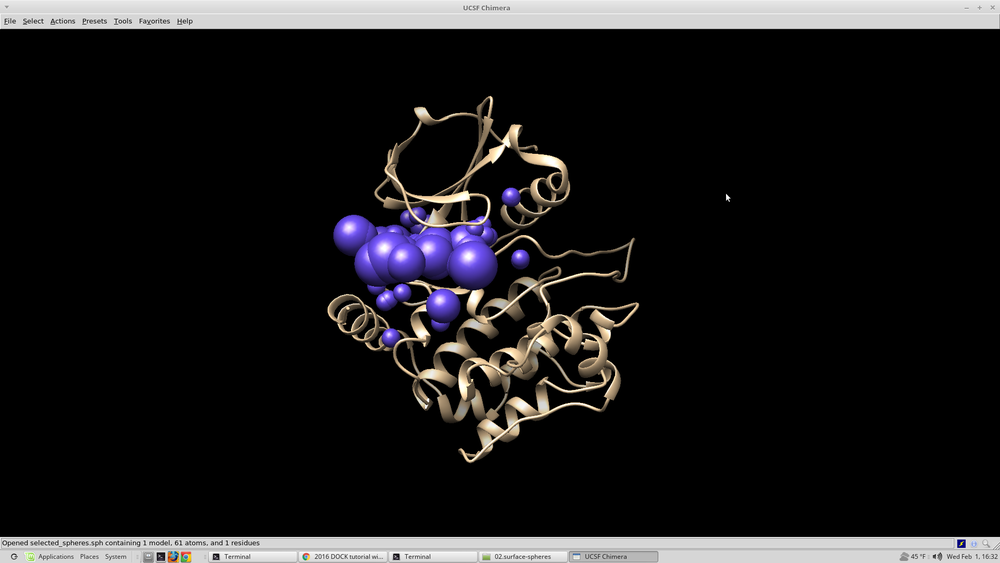
To visualize the spheres using Chimera as previously done: Launch Chimera, choose File -> Open, choose 4qmz.rec.noH.pdb File -> Open, choose output_spheres_selected.pdb Select -> Residue -> SPH Actions -> Atoms/Bonds -> sphere The selected spheres with the receptor surface should look similar to that as seen below:
IV. Generating Box and Grid
enter directory 03.box-grid
create input showbox.in
Y
8.0
../02.surface-spheres/selected_spheres.sph
1
4qmz.box.pdb
This input designates to dock that:
we want to create a box, the box length should be 8.0 Angstroms, use the selected spheres in the file designated, output the box to the file specified
to use this input type:
showbox < showbox.in
This box can be visualized in chimera using a similar approach as visualizing the spheres
Compute the energy grid
create grid.in file
vi grid.in
grid.in needs to contain:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 12 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../01.dockprep/4qmz.rec.mol2 box_file ../03.box-grid/4qmz.box.pdb vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
this script should output verbosely to the terminal and produce 2 output files, grid.bmp and grid.nrg, both binary files.
grid -i grid.in > gridinfo.out
Check the standard output file gridinfo.out to ensure that individual charges on charged residues are integer values (-1 or +1).
University of California at San Francisco, DOCK 4.0.1
__________________Job_Information_________________
launch_time Wed Feb 8 17:05:50 2017
host_name unknown
memory_limit -1
working_directory /home/campus.stonybrook.edu/ronassar/MySBFiles/projects/dock_tutorial/03.box-grid2_savegridoutput
user_name ronassar
________________General_Parameters________________
compute_grids yes
grid_spacing 0.4
output_molecule no
________________Scoring_Parameters________________
contact_score no
energy_score yes
energy_cutoff_distance 9999
atom_model a
attractive_exponent 6
repulsive_exponent 12
distance_dielectric yes
dielectric_factor 4
bump_filter yes
bump_overlap 0.75
____________________File_Input____________________
receptor_file ../01.dockprep/4qmz.rec.mol2
box_file ../03.box-grid/4qmz.box.pdb
vdw_definition_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn
____________________File_Output___________________
score_grid_prefix grid
Reading in coordinates of receptor.
CHARGED RESIDUE MET : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE ASP : -1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE ARG : 1.000
CHARGED RESIDUE LYS : 1.000
CHARGED RESIDUE GLU : -1.000
CHARGED RESIDUE SER : -1.000
Total charge on 4qmz_rec_leap.pdb : -5.000
Reading in grid box information.
Box center of mass : -1.912 0.054 14.111
Box dimensions : 39.856 41.687 38.838
Number of grid points per side [x y z] : 101 106 99
Total number of grid points : 1059894
Generating scoring grids.
Percent of protein atoms processed : 0
Percent of protein atoms processed : 10
Percent of protein atoms processed : 20
Percent of protein atoms processed : 30
Percent of protein atoms processed : 40
Percent of protein atoms processed : 50
Percent of protein atoms processed : 60
Percent of protein atoms processed : 70
Percent of protein atoms processed : 80
Percent of protein atoms processed : 90
Percent of protein atoms processed : 100
Writing general grid info to grid.bmp
Writing bump grid to grid.bmp
Writing energy grids to grid.nrg
Writing attractive VDW energy grid
Writing repulsive VDW energy grid
Writing electrostatic energy grid
Finished calculation.
V. Docking a Single Molecule for Pose Reproduction
Create or enter directory 4, 04.dock
Minimization
create min.in
vi min.in
or
touch min.in
min.in should contain:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01.dockprep/4qmz.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
run:
dock6 -i min.in
this command will output 4qmz.lig.min_scored.mol2
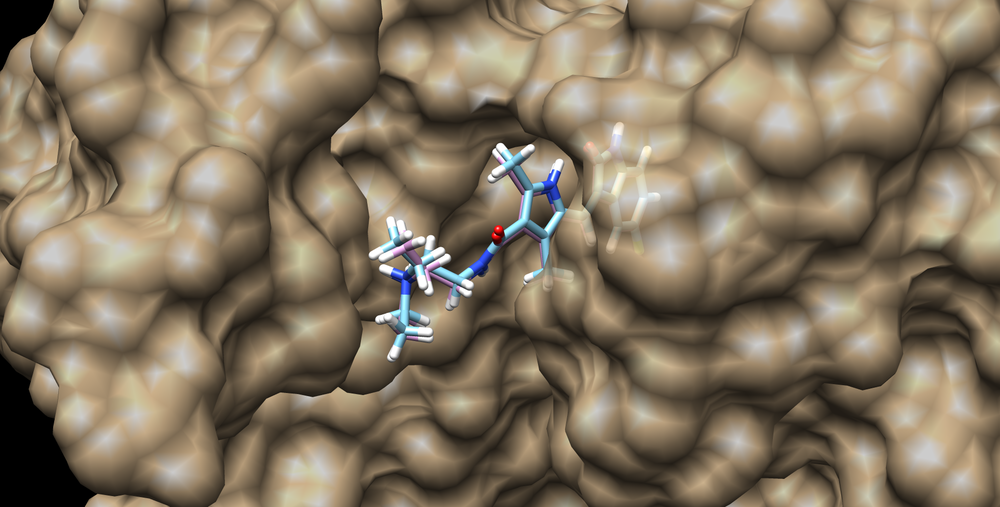
This structure can be visualized using chimera and loading the surface into chimera along with the minimized and unminimized ligand structures:
The RMSD of the minimized ligand to crystal structure ligand is 0.297 A.
At this point we are going to calculate the van der waals and electrostatic footprints for the unminimized and minimized ligand structures in relation to the receptor active site.
create the input file: footprint.in
conformer_search_type rigid use_internal_energy no ligand_atom_file ./4qmz.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_use_footprint_reference_mol2 yes fps_footprint_reference_mol2_filename ../01.dockprep/4qmz.lig.mol2 fps_foot_compare_type Euclidean fps_normalize_foot no fps_foot_comp_all_residue yes fps_receptor_filename ../01.dockprep/4qmz.rec.mol2 fps_vdw_att_exp 6 fps_vdw_rep_exp 12 fps_vdw_rep_rad_scale 1 fps_use_distance_dependent_dielectric yes fps_dielectric 4.0 fps_vdw_fp_scale 1 fps_es_fp_scale 1 fps_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix fps.min.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
this footprint calculation can be run in the same way as the minimization:
dock6 -i footprint.in
This will output three files: fps.min.output_scored.mol2
fps.min.output_scored_footprint_scored.txt
fps.min.output_hbond_scored.txt
With an in house script, plot_footprint_single_magnitude.py the two sets of footprints will be plotted for comparison
this script will be run using:
python plot_footprint_single_magnitude.py fps.min.output_scored_footprint_scored.txt 50
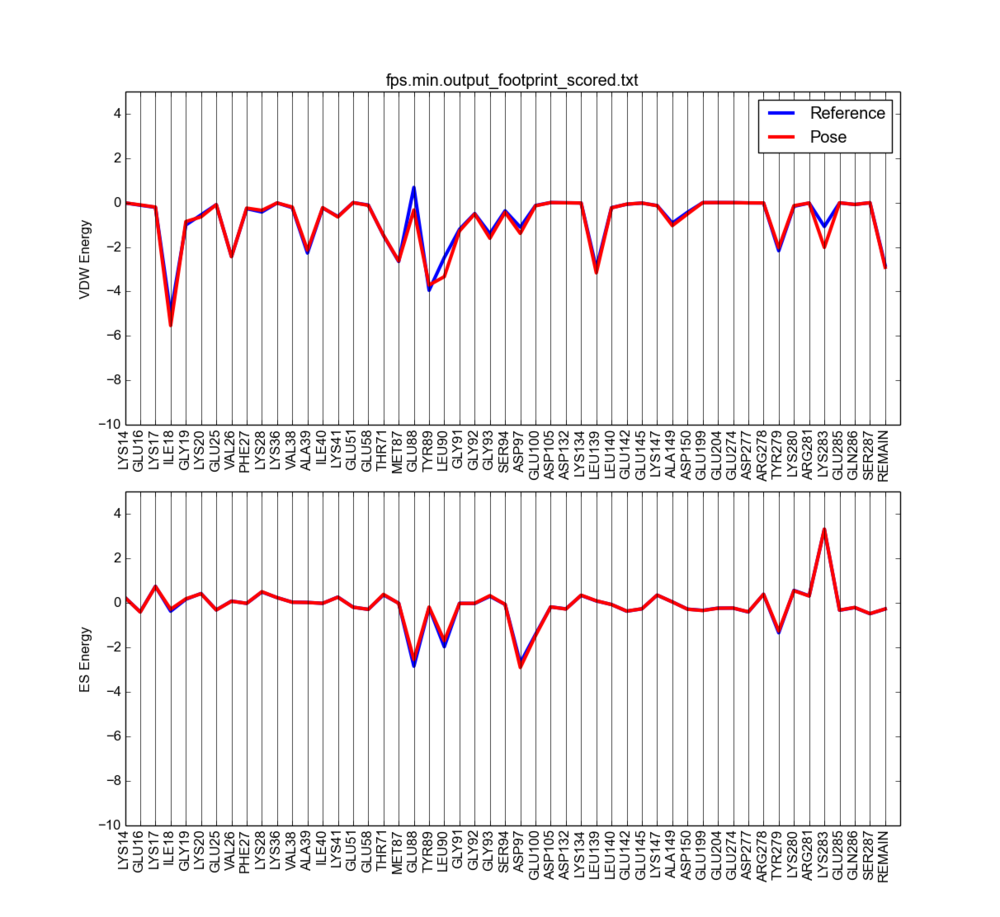
The output will look something like this:
Notice the vdW clashes are now reduced (vdW positive peaks disappeared).
Rigid Docking
To help identify any errors in preparing the ligand and receptor files, a rigid docking, fixed growth docking and flexible docking runs will be performed. This is a check that the program can put the ligand into the crystal structure pose before proceeding.
Create an input file 'rigid.in' for the rigid docking run with the following parameters:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01.dockprep/4qmz.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.rigid_norestraint write_orientations no num_scored_conformers 50 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Outputs the file "4qmz.rigid_norestraint" the no restraint refers to the option of the parameter simplex_restraint_min in the input file. This removes the tether when doing the minimization. The best scoring structure had an RMSD of 1.??? to the native ligand pose.
Fixed Anchor Docking
In fixed anchor docking, the ligand location is fixed in the active site and the rotatable bonds space is searched (i.e angles are flexible).
Create an input file 'fad.in' for the rigid docking run with the following parameters: Notice the search algorithm is now rigidset to flex (for flexible) and orient option is set to no (for fixing the anchor in place).
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01.dockprep/4qmz.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand no automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.fad write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Flexible Docking
In flexible docking, the ligand location and rotatable bonds are search.
Create an input file 'flex.in' for the flexible docking run with the following parameters: Notice the search algorithm is now set to flex (for flexible) and orient option is set to yes (for fixing the anchor in place).
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01.dockprep/4qmz.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /opt/AMS536/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /opt/AMS536/dock6/parameters/flex.defn flex_drive_file /opt/AMS536/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.flex write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
VI. Virtual Screening
At this point we will begin docking potential inhibitors into the active site of 4qmz. The compound library we will use contains 25,000 molecules and was previously prepared for another docking experiment. The compound library contains molecules with rotatable bonds number between 7 and 15. The library used is titled small_ligand_library.mol2, this library can be found at /gpfs/projects/AMS536/virtual_screen_library on seawulf.
We began by copying all of our local directories outlined in the first section to a project directory on seawulf to allow use to expedite the docking process over the 25,000 compounds in our test library.
We will create a virtual-screen.in file that looks like the following:
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file small_ligand_library.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/4qmz.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.virtualscreen write_orientations no num_scored_conformers 1 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
This script is submitted to the seawulf cluster using a script called screen.sh which contains the following:
#! /bin/tcsh #PBS -l nodes=4:ppn=28 #PBS -l walltime=24:00:00 #PBS -o jobstatus.out #PBS -e joberror.err #PBS -N large_vs #PBS -V #PBS -q long cd $PBS_O_WORKDIR mpirun -np 112 dock6.mpi -i virtual-screen.in -o virtualscreen1.out
This screen.sh input specifies that we will use 112 processors to dock our virtual library. We have a wall clock for the job of 24 hours and we will output jobstatus.out, joberror.err and virtualscreen1.out. In addition to these output files there is an output for the work performed by each processor individually.
Cartesian Minimization of the docked molecules
Now we will do cartesian minimization of the docked molecules. Cartesian here is referring to the receptor being explicitly present during the minimization.
Write the minimization input script minimize-vs.in:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 4qmz.virtualscreen_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../01.dockprep/4qmz.rec.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix 4qmz.virtualscreen1.minimized write_orientations no num_scored_conformers 1 rank_ligands no
Submit the minimization job using with the script "job-min.sh" following lines:
#!/bin/tcsh #PBS -l nodes=16:ppn=28 #PBS -l walltime=12:00:00 #PBS -o jobstatusmin.out #PBS -e jobstatusmin.err #PBS -N chunk0min #PBS -V #PBS -q medium module load intel/mpi/64/2017/0.098 cd $PBS_O_WORKDIR mpirun -np 448 /gpfs/projects/AMS536/zzz.programs/dock6/bin/dock6.mpi -i minimize-vs.in -o minimize-vs.out
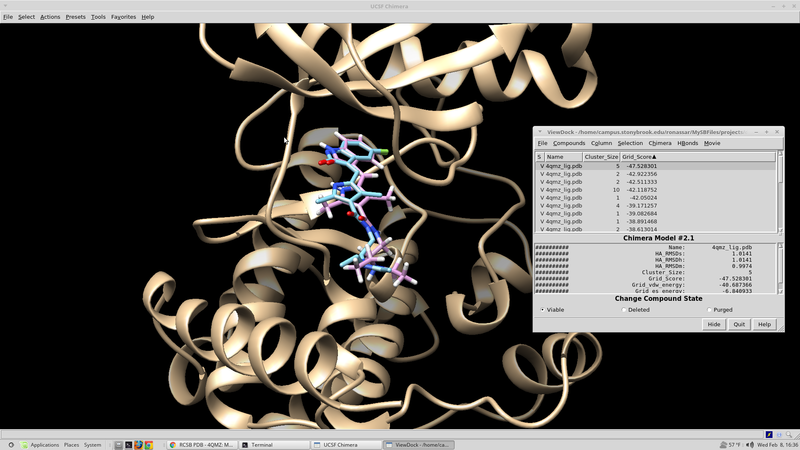
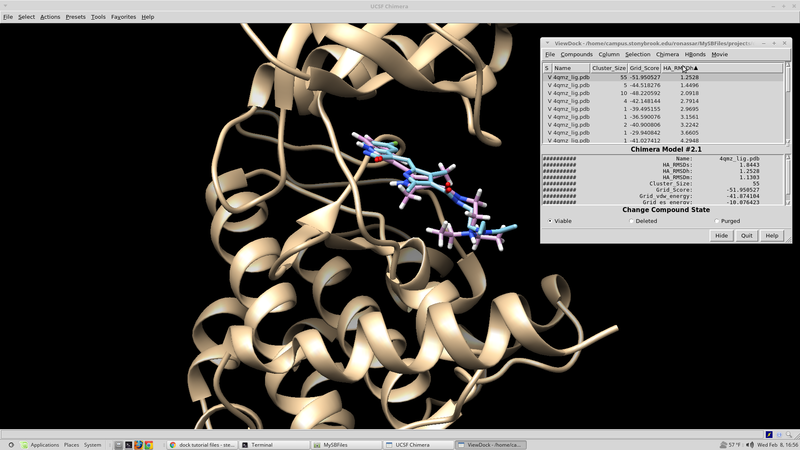
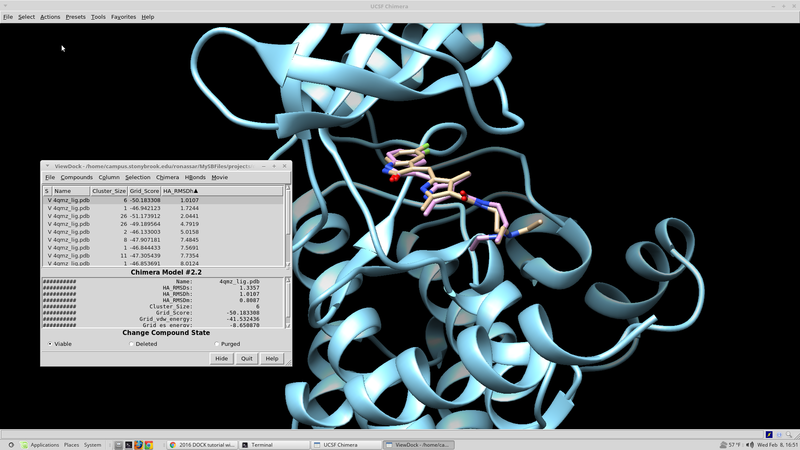
At this point it would be good to rank our docked ligands by score and extract the 100 best scoring (100 lowest scoring, most negative) ligands to investigate further.
Rescoring Docked Molecules
Dock has several built in scoring functions. In an effort to utilize these alternative scoring functions it is worthwhile to rescore the results from the initial virtual screen that were collected. The virtual screen will be rescored using footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score. Each of these scoring functions focuses in on a different chemical aspect of the ligand in question. The rescore.in script for this run can be found here:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../05.virtual- screen/4qmz.virtualscreen1.minimized_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_energy no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_use_footprint_reference_mol2 yes descriptor_fps_footprint_reference_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 descriptor_fps_foot_compare_type Euclidean descriptor_fps_normalize_foot no descriptor_fps_foot_comp_all_residue yes descriptor_fps_receptor_filename ../01.dockprep/4qmz.rec.mol2 descriptor_fps_vdw_att_exp 6 descriptor_fps_vdw_rep_exp 12 descriptor_fps_vdw_rep_rad_scale 1 descriptor_fps_use_distance_dependent_dielectric yes descriptor_fps_dielectric 4.0 descriptor_fps_vdw_fp_scale 1 descriptor_fps_es_fp_scale 1 descriptor_fps_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../04.dock/4qmz.lig.min_scored.mol2 descriptor_hungarian_ref_filename ../04.dock/4qmz.lig.min_scored.mol2 descriptor_hungarian_matching_coeff -5 descriptor_hungarian_rmsd_coeff 1 descriptor_volume_reference_mol2_filename ../04.dock/4qmz.lig.min_scored.mol2 descriptor_volume_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hungarian 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
From here the results should be sorted according the score based on the scoring function in question. A script for this can be written fairly quickly in whatever language you are most comfortable with. If you are not very experienced with computer programming, a python script developed in house can be found here that will search through the output .mol2 file and select the top N scoring molecules. These top scoring molecule s are taken from the virtual screen "top'N'_molid.txt" and finds and their coordinates saved to an output file "top'N'_molid.mol2".
rank_scores_get_coords.py:
#!/usr/bin/python
import sys
from operator import itemgetter, attrgetter, methodcaller
# input mol2 file as command line argument
f0=open(sys.argv[1],'r') ; data0 = f0.readlines() ; f0.close() #open mol2 data file with scored molecules
# define variables
nummol = 50 # number of molecules to keep after ranking by score
#scoretype="Grid_Score:" # scoring function to rank by
scoretype="Footprint_Similarity_Score:"
#--------------------------------------------------------------------------------------------------------------------------
list_molid=[] #create a list to store molecule ids
list_molscores=[] #create list to store scores for each respective molecule
for line in data0: #reads through the input file
if line != "\n": #if the line is not empty continue
if line.split()[0]=="##########": #if the line starts with ############ then continue
if line.split()[1]=="Name:": #if the line has the next entry as Name: continue
molid = line.split()[2] #then add the molID to list_molid
list_molid.append(molid)
elif line.split()[1]==scoretype:
molscore = line.split()[2]
list_molscores.append(molscore)
#print list_molid
#print list_molscores
both = zip(list_molid,list_molscores) #concatenates list_molid and list_molscores into one array
sorted_both=sorted(both, key=itemgetter(1), reverse=True) #sorts the "both" list
sorted_top=[] #names a new list sorted_top5
#for element in sorted_both: #print out first n entries in sorted_both
# print element[0], element[1]
o=open("top"+str(nummol)+"_molid.txt","w") #opens output file topx_molid.txt
for element in sorted_both[:nummol]:
sorted_top.append(element[0])
o.write(element[0]+"\n") #stores the top nummol entries in sorted_top
o.close()
#print sorted_top
##
##--------------------------------------------------------------------------------------------------------------------------
##
## Get coordinates of molecules of interest from scored mol2
f1=open("top"+str(nummol)+"_molid.txt","r") ; data1=f1.readlines(); f1.close() # takes in sorted list of top molecules
f2=open(sys.argv[1],"r") ; data2=f2.readlines(); f2.close() # takes in mol2 results
identifier=0 # this will switch between 0 and 1. When it is 0 no line printing, when it is 1 we print the line.
molid_top=[] # list of molecule ids that are of interest
# go through the first file (has top n molecules) and append those ids to molecule ids list
for line in data1:
molid=line.split()[0]
molid_top.append(molid)
output=open("top"+str(nummol)+"_molid.mol2","w")
# go through the second file (the rescored mol2 file), the next commands print out the coordinates for the molecules of interest
for line in data2:
# if line != "\n": # skipping empty lines
if identifier==1:
try:
if line.split()[0]=="##########" and line.split()[1]=="Name:" and line.split()[2] not in molid_top: # changing the identifier when hitting a new molecule that is not of interest
identifier=0
output.write('\n')
else:
output.write(line) # else keep the identifier at 1 and keep printing
except IndexError:
continue
try:
if identifier==0 and line.split()[0]=="##########" and line.split()[1]=="Name:" and line.split()[2] in molid_top: # change identifier to 1 when we hit molecule of interest
identifier=1
output.write(line)
except IndexError:
continue
output.close()
VIII. Frequently Encountered Problems
Spending the time to ensure your starting structure is reasonable is of the utmost importance and will save you much headache moving forward in your study. Reading through the PDB file as downloaded from the protein data bank will give the researcher a lot of information regarding issues or grey area encountered by the researchers responsible for publishing the structure initially. Threading a structure through some quality control software such as tleap, chimera or another comparable software package will ensure charges are reasonable, hydrogens are appropriately placed, and there are no missing residues in the peptide chain.
A basic understanding of your enzymes active site will also be beneficial while approaching an enzymatic system from a docking perspective. Some questions to ask oneself are: does my protein have an ion (Mg, Ca, etc.) in the active site? Does my ligand make important interactions with conserved water residues? If so these waters could be kept in with the receptor to aid in discovery. The alternative would be to design an inhibitor that displaces the water molecules and supplements favorable interactions mediated by the water between the ligand and protein.