Difference between revisions of "2020 DOCK tutorial 3 with PDBID 4F4P"
Stonybrook (talk | contribs) (→Flexible Docking) |
Stonybrook (talk | contribs) (→IX Rescoring Docked Molecules) |
||
| (45 intermediate revisions by the same user not shown) | |||
| Line 31: | Line 31: | ||
===Prepare the Receptor file=== | ===Prepare the Receptor file=== | ||
| − | With the PDB file loaded by chimera, do Select -> Residue -> SO4 to select the sulfate. Use Actions -> Atoms/Bonds -> Delete to delete it. Then do Select -> Residue -> HOH to select the water molecules. Use the same method to delete it. Then do Select -> Structure -> (0SB) to select ligand molecule. Use the same method to delete it. Save the receptor as 4f4p_rec_noh.mol2. | + | With the PDB file loaded by chimera, do |
| + | Select -> Residue -> SO4 | ||
| + | to select the sulfate. Use | ||
| + | Actions -> Atoms/Bonds -> Delete | ||
| + | to delete it. Then do | ||
| + | Select -> Residue -> HOH | ||
| + | to select the water molecules. Use the same method to delete it. Then do | ||
| + | Select -> Structure -> (0SB) | ||
| + | to select ligand molecule. Use the same method to delete it. Save the receptor as 4f4p_rec_noh.mol2. | ||
OR | OR | ||
| − | With the PDB file loaded by chimera, hold the control button and click on the protein chain to select a protein residue. Then click up arrow button to select the whole protein chain (Receptor). Then Select -> Invert(all models) to invert your selection (This will select all the components in your PDB file except receptor). Then do Actions->Atoms/bonds->Delete. You will remain with a protein receptor. Save the receptor as 4f4p_rec_noh.mol2. | + | With the PDB file loaded by chimera, hold the control button and click on the protein chain to select a protein residue. Then click up arrow button to select the whole protein chain (Receptor). Then |
| + | Select -> Invert(all models) | ||
| + | to invert your selection (This will select all the components in your PDB file except receptor). Then do | ||
| + | Actions->Atoms/bonds->Delete. | ||
| + | You will remain with a protein receptor. Save the receptor as 4f4p_rec_noh.mol2. | ||
| Line 42: | Line 54: | ||
===Prepare the Ligand File=== | ===Prepare the Ligand File=== | ||
Open the pdf file again. | Open the pdf file again. | ||
| − | Do Select -> Structure -> ligand(0SB) to select the ligand. Do Select -> Invert (all models), then delete the selected atoms. This will left the ligand molecule only. Save it as 4f4p_lig_noh.mol2. | + | Do |
| + | Select -> Structure -> ligand(0SB) | ||
| + | to select the ligand. Do | ||
| + | Select -> Invert (all models), | ||
| + | then delete the selected atoms. This will left the ligand molecule only. Save it as 4f4p_lig_noh.mol2. | ||
[[File:4f4p_lig_noh.png|thumb|center|500px|Figure 3: Ligand with no Hydrogens and charges (4f4p_lig_noh.mol2)]] | [[File:4f4p_lig_noh.png|thumb|center|500px|Figure 3: Ligand with no Hydrogens and charges (4f4p_lig_noh.mol2)]] | ||
| Line 90: | Line 106: | ||
sphgen -i INSPH -o OUTSPH | sphgen -i INSPH -o OUTSPH | ||
| − | Once sphgen command is successful, | + | Once sphgen command is successful, following output files will be created. |
| + | OUTSPH | ||
| + | 4f4p_rec.sph | ||
| + | |||
| + | Open the f4p_rec.sph using Chimera along with 4f4p_rec_noh.mol2 file. You should get a similar output like the image below. | ||
| + | |||
[[File:4f4precnohsph.png|thumb|center|700px|Figure 6:Chimera view of generated spheres along with the receptor(4f4p_rec_noh.mol2 + 4f4p_rec.sph)]] | [[File:4f4precnohsph.png|thumb|center|700px|Figure 6:Chimera view of generated spheres along with the receptor(4f4p_rec_noh.mol2 + 4f4p_rec.sph)]] | ||
| Line 99: | Line 120: | ||
sphere_selector 4f4p_rec.sph ../001.files/4f4p_noh_lig.mol2 10.0 | sphere_selector 4f4p_rec.sph ../001.files/4f4p_noh_lig.mol2 10.0 | ||
| − | This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph. Visualize the selected spheres using Chimera to make sure the correct spheres are selected. Notice that, spheres around the ligand binding site are kept and all the other spheres are deleted | + | This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph. |
| + | |||
| + | Output files: | ||
| + | selected_spheres.sph | ||
| + | |||
| + | Visualize the selected spheres using Chimera to make sure the correct spheres are selected. Notice that, spheres around the ligand binding site are kept and all the other spheres are deleted by following sphere selector command . | ||
[[File: 4f4pre.png |thumb|center|700px|Figure 7: Selected spheres along with the receptor (4f4p_rec_noh.mol2 + selected_spheres.sph)]] | [[File: 4f4pre.png |thumb|center|700px|Figure 7: Selected spheres along with the receptor (4f4p_rec_noh.mol2 + selected_spheres.sph)]] | ||
| Line 119: | Line 145: | ||
If this step is successful, you should see a new file (4f4p.box.pdb) in 003.gridbox folder. | If this step is successful, you should see a new file (4f4p.box.pdb) in 003.gridbox folder. | ||
| + | |||
| + | Output file: | ||
| + | 4f4p.box.pdb | ||
[[File:4f4p_rec_noh_selected_sphere_box.png|thumb|center|500px|Figure 8:surface of the receptor,selected spheres with box]] | [[File:4f4p_rec_noh_selected_sphere_box.png|thumb|center|500px|Figure 8:surface of the receptor,selected spheres with box]] | ||
| Line 146: | Line 175: | ||
grid -i grid.in -o grid.out | grid -i grid.in -o grid.out | ||
| + | |||
| + | If this is successful, you will get following output files. | ||
| + | |||
| + | grid.out | ||
| + | grid.nrg | ||
| + | grid.bmp | ||
=V. Docking= | =V. Docking= | ||
| Line 207: | Line 242: | ||
rank_ligands no | rank_ligands no | ||
| − | Save the file and run the following command | + | Save the file and run the following command. |
dock6 -i min.in -o min.out | dock6 -i min.in -o min.out | ||
| + | |||
| + | If you have done correctly, you will get followig out files. | ||
| + | |||
| + | min.out | ||
| + | 4f4p.lig.min_scored.mol2 | ||
[[File:Rec min lig complex.png |thumb|center|700px|Figure 9: Receptor with energy minimized ligand]] | [[File:Rec min lig complex.png |thumb|center|700px|Figure 9: Receptor with energy minimized ligand]] | ||
| Line 272: | Line 312: | ||
rank_ligands no | rank_ligands no | ||
| − | If | + | If footprint runs successfully, you can find the following files generated |
| + | footprint.out | ||
fps.min.output_footprint_scored.txt | fps.min.output_footprint_scored.txt | ||
fps.min.output_hbond_scored.txt | fps.min.output_hbond_scored.txt | ||
| + | fps.min.output_scored.mol2 | ||
| + | |||
In order to visualize the footprint, first we need to copy a python script into our directory, then run it: | In order to visualize the footprint, first we need to copy a python script into our directory, then run it: | ||
| Line 353: | Line 396: | ||
Run dock using the created input file. | Run dock using the created input file. | ||
| − | dock6 - | + | dock6 -i rigid.in -o rigid.out |
| + | |||
| + | Once rigid docking is successful, you will get following output files. | ||
| + | |||
| + | rigid.out | ||
| + | rigid.out_conformers.mol2 | ||
| + | rigid.out_scored.mol2 | ||
| + | |||
| + | In order to check the rigid docking success, open rigid.out_scored.mol2 output file using Chimera by following steps. | ||
| + | |||
| + | Open Chimera | ||
| + | File -> Open -> 4f4p_rec_h.mol2 (Actions -> Surface -> Show) | ||
| + | File -> Open -> 4f4p_lig_h.mol2 | ||
| + | Tools -> Surface/binding Analysis -> ViewDock -> Select the Rigid Dock output file. (rigid.out_scored.mol2) | ||
| + | In the loaded dialog box select Dock4,5 or 6 | ||
| + | |||
| + | Once everything is loaded go to the ViewDock window and use it's menu to view all the calculated properties regarding the rigid docked ligand by following the steps below. | ||
| + | |||
| + | Column -> Show -> Grid_Score | ||
| + | Column -> Show -> HA_RMSDs | ||
| + | |||
| + | Follow the same steps to get all different properties as you interested. | ||
| + | |||
| + | You can choose different conformations from the ViewDock window. If you sorted it, the top one should be the conformation with lowest energy. Your visualized structure should be similar to the image below. | ||
| − | |||
[[File:rigid_out1.png|thumb|center|500px|Figure 11: rigid out file (different poses with their corresponding grid scores and RMSDs are listed in the table (left), best confirmation with lowest RMSD value (cyan) is shown compared to the original ligand)]] | [[File:rigid_out1.png|thumb|center|500px|Figure 11: rigid out file (different poses with their corresponding grid scores and RMSDs are listed in the table (left), best confirmation with lowest RMSD value (cyan) is shown compared to the original ligand)]] | ||
| Line 364: | Line 429: | ||
Create an input file called fad.in and use the following command. | Create an input file called fad.in and use the following command. | ||
| − | dock6 -i fad.in | + | dock6 -i fad.in -o fad.out |
Use the following lines to answer the prompted questions as we did in rigid docking. | Use the following lines to answer the prompted questions as we did in rigid docking. | ||
| Line 446: | Line 511: | ||
rank_ligands no | rank_ligands no | ||
| − | Once docking is completed | + | Once docking is completed following output files will be generated. |
| + | |||
| + | fad.out | ||
| + | 4f4p_fad_scored.mol2 | ||
| + | |||
| + | Follow the same method used in rigid docking to visualize the docked poses using Chimera by opening the 4f4p_fad_scored.mol2 file. Once it is visualized, it should like the image below. | ||
[[File:Fad_out.png|thumb|center|500px|Figure 13: Fixed Anchor docking out file (different poses with their corresponding grid scores and RMSDs are listed in the table (left), best confirmation with lowest RMSD value (cyan) is shown compared to the original ligand)]] | [[File:Fad_out.png|thumb|center|500px|Figure 13: Fixed Anchor docking out file (different poses with their corresponding grid scores and RMSDs are listed in the table (left), best confirmation with lowest RMSD value (cyan) is shown compared to the original ligand)]] | ||
| Line 456: | Line 526: | ||
Create a new input file for flexible docking called flex.in and perform flexible docking using DOCK6 by following command. | Create a new input file for flexible docking called flex.in and perform flexible docking using DOCK6 by following command. | ||
| − | dock6 -i flex.in | + | dock6 -i flex.in -o flex.out |
Answer the prompted questions using the following lines as we did in rigid and fixed anchor docking (or copy the following content in to flex.in file and save). | Answer the prompted questions using the following lines as we did in rigid and fixed anchor docking (or copy the following content in to flex.in file and save). | ||
| Line 475: | Line 545: | ||
internal_energy_rep_exp 12 | internal_energy_rep_exp 12 | ||
internal_energy_cutoff 100.0 | internal_energy_cutoff 100.0 | ||
| − | ligand_atom_file 4f4p.lig.min_scored.mol2 | + | ligand_atom_file ../004.dock/4f4p.lig.min_scored.mol2 |
limit_max_ligands no | limit_max_ligands no | ||
skip_molecule no | skip_molecule no | ||
| Line 481: | Line 551: | ||
calculate_rmsd yes | calculate_rmsd yes | ||
use_rmsd_reference_mol yes | use_rmsd_reference_mol yes | ||
| − | rmsd_reference_filename 4f4p.lig.min_scored.mol2 | + | rmsd_reference_filename ../004.dock/4f4p.lig.min_scored.mol2 |
use_database_filter no | use_database_filter no | ||
orient_ligand yes | orient_ligand yes | ||
| Line 499: | Line 569: | ||
grid_score_vdw_scale 1 | grid_score_vdw_scale 1 | ||
grid_score_es_scale 1 | grid_score_es_scale 1 | ||
| − | grid_score_grid_prefix ../003. | + | grid_score_grid_prefix ../003.gridbox/grid |
multigrid_score_secondary no | multigrid_score_secondary no | ||
dock3.5_score_secondary no | dock3.5_score_secondary no | ||
| Line 534: | Line 604: | ||
rank_ligands no | rank_ligands no | ||
| − | Once flexible docking is completed | + | Once flexible docking is completed following output files will be generated. |
| + | |||
| + | flex.out | ||
| + | flex.out_scored.mol2 | ||
| + | |||
| + | Use the visualization steps used in rigid and fixed anchor docking and study the properties of the docking results by opening flex.out_scored.mol2 file. (You will get one confirmation since you're writing num_scored_conformers as one in the above script.) | ||
| + | |||
| + | [[File:Flex_out.png|thumb|center|500px|Figure 15: Flexible docking out file (pose with corresponding grid score and RMSD are listed in the table (As we got one confirmation after flexible docking) (left), confirmation with its RMSD value (cyan) is shown compared to the original ligand)]] | ||
| + | |||
| + | = VI. Virtual Screen = | ||
| + | Virtual screening is the method of screening a ligand library (drug-like molecules) to filter the best ligands which can bind to the binding site of a specific receptor. Here we will be using a ligand library which contains 25000 molecules to select the best ligands which can be used to replace the original ligand that the PDB file contained. | ||
| + | |||
| + | Move to the directory 005.virtual_screen. | ||
| + | |||
| + | Copy the ligand library to the same directory. | ||
| + | |||
| + | cp /gpfs/projects/AMS536/2020/536_class/VS_library_5K.mol2 ./ | ||
| + | |||
| + | Create a new input file for virtual screen. | ||
| + | |||
| + | touch virtual.in | ||
| + | |||
| + | Use the input file to perform virtual screen using DOCK6. | ||
| + | |||
| + | dock6 -i virtual.in -o virtual.out | ||
| + | |||
| + | Use the following lines to answer the prompted questions. | ||
| + | |||
| + | conformer_search_type flex | ||
| + | write_fragment_libraries no | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 9 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file VS_library_5K.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../002.surface_spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../003.gridbox/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix virtual.out | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | You can run this job on head node for a short while, just to make sure everything is fine. Then you can kill the job by Ctrl-C. | ||
| + | |||
| + | = VII Virtual Screeni mpi = | ||
| + | |||
| + | Go to 006. virtual_screen_mpi directory and copy the virtual.in file here. We will run the job on the seawulf cluster using slurm script. | ||
| + | |||
| + | Creat the virtual.sh file | ||
| + | vi virtual.sh | ||
| + | |||
| + | Add the following lines there | ||
| + | |||
| + | #!/bin/sh | ||
| + | #SBATCH --partition=long-40core | ||
| + | #SBATCH --time=48:00:00 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --ntasks=1 | ||
| + | #SBATCH --job-name=........#whatever you want | ||
| + | #SBATCH --output=%x-%j.o | ||
| + | |||
| + | dock6 -i virtual.in -o virtual.out | ||
| + | |||
| + | Submit the job on seawulf using | ||
| + | sbatch virtual.sh | ||
| + | To check the status of your job | ||
| + | squeue -u [NETID] | ||
| + | |||
| + | ===Check Virtual Screen output=== | ||
| + | |||
| + | The job was terminated due to time limit. In order to calculate how many molecules were screened, we run the command: | ||
| + | grep "Molecule: ZINC" virtual.out | wc -l | ||
| + | |||
| + | Everyone in our group has got either 1066, 1068, 1061 molecules processed. | ||
| + | |||
| + | To vizualize the output, we open the file virtual.out_scored.mol2 with Chimera. The Chimera could be slow as there are many many molecules. | ||
| + | Go to Tools -> Surface binding Analysis -> View Dock. Also open the receptor in this session. | ||
| + | |||
| + | Column -> Show -> Grid Score | ||
| + | |||
| + | Column -> Show -> Mass weight | ||
| + | |||
| + | We see that best Grid scored molecules have higher energy than those that have lower Grid score energy. | ||
| + | The vdw energy also have the same trend with Grid score. | ||
| + | |||
| + | =VIII.Cartesian Minimization= | ||
| + | |||
| + | Here we will use the docked molecules and perform a cartesian minimization of them. | ||
| + | |||
| + | Move to the directory 007.cartesianmin. | ||
| + | |||
| + | Create a new input file for the minimization. | ||
| + | touch min.in | ||
| + | Use the input file to perform the cartesian minimization using DOCK6. | ||
| + | dock6 -i min.in -o min.out | ||
| + | |||
| + | Use the following lines to answer the prompted questions. | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../005.virtual_screen/virtual.out_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary yes | ||
| + | continuous_score_secondary no | ||
| + | cont_score_rec_filename ../001.files/4f4p_rec_h.mol2 | ||
| + | cont_score_att_exp 6 | ||
| + | cont_score_rep_exp 12 | ||
| + | cont_score_rep_rad_scale 1 | ||
| + | cont_score_use_dist_dep_dielectric yes | ||
| + | cont_score_dielectric 4.0 | ||
| + | cont_score_vdw_scale 1 | ||
| + | cont_score_es_scale 1 | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix 4f4p.virtualscreen.minimized | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Then in order to submit the script in to seawulf cluster, open a new script called cartesianmin.sh and write the following. | ||
| + | |||
| + | #!/bin/sh | ||
| + | #SBATCH --partition=long-40core | ||
| + | #SBATCH --time=48:00:00 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --ntasks=1 | ||
| + | #SBATCH --job-name=4f4p_cartesian | ||
| + | #SBATCH --output=%x-%j.o | ||
| + | |||
| + | echo "============================= SLURM JOB =================================" | ||
| + | date | ||
| + | echo | ||
| + | echo " The job will be started on the following node(s):" | ||
| + | echo $SLURM_JOB_NODELIST | ||
| + | echo | ||
| + | echo "Slurm user: $SLURM_JOB_USER" | ||
| + | echo "Run directory: $(pwd)" | ||
| + | echo "Job ID: $SLURM_JOB_ID" | ||
| + | echo "Job name: $SLURM_JOB_NAME" | ||
| + | echo "Partition: $SLURM_JOB_PARTITION" | ||
| + | echo "Number of nodes: $SLURM_JOB_NUM_NODES" | ||
| + | echo "Number of tasks: $SLURM_NTASKS" | ||
| + | echo "Submitted from: $SLURM_SUBMIT_HOST:$SLURM_SUBMIT_DIR" | ||
| + | echo "=========================================================================" | ||
| + | |||
| + | dock6 -i min.in -o min.out | ||
| + | |||
| + | Then submit the job into seawulf cluster. | ||
| + | sbatch cartesianmin.sh | ||
| + | |||
| + | Once it has done correctly, you will get a output file called 4f4p.virtualscreen.minimized_scored.mol2. This file contains molecules with minimized energies by removing unfavorable steric clashes. | ||
| + | |||
| + | = IX Rescoring Docked Molecules = | ||
| + | |||
| + | In order to rank our docked ligands and extract the best ligands (which has the most negative, lowest scores) based on different scoring functions like Footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score, we will be using Dock6 to rescore the virtual screen minimized ligands. | ||
| + | |||
| + | Move to the directory 008.rescore and create a new input file for the rescoring. | ||
| + | touch rescore.in | ||
| + | |||
| + | Use the input file to perform the cartesian minimization using DOCK6. | ||
| + | |||
| + | dock6 -i rescore.in | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../007.cartesianmin/4f4p.virtualscreen.minimized_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score no | ||
| + | descriptor_use_multigrid_score no | ||
| + | descriptor_use_continuous_score yes | ||
| + | descriptor_use_footprint_similarity yes | ||
| + | descriptor_use_pharmacophore_score yes | ||
| + | descriptor_use_tanimoto yes | ||
| + | descriptor_use_hungarian yes | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_cont_score_rec_filename ../001.files/4f4p_rec_h.mol2 | ||
| + | descriptor_cont_score_att_exp 6 | ||
| + | descriptor_cont_score_rep_exp 12 | ||
| + | descriptor_cont_score_rep_rad_scale 1 | ||
| + | descriptor_cont_score_use_dist_dep_dielectric yes | ||
| + | descriptor_cont_score_dielectric 4.0 | ||
| + | descriptor_cont_score_vdw_scale 1 | ||
| + | descriptor_cont_score_es_scale 1 | ||
| + | descriptor_fps_score_use_footprint_reference_mol2 yes | ||
| + | descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 | ||
| + | descriptor_fps_score_foot_compare_type Euclidean | ||
| + | descriptor_fps_score_normalize_foot no | ||
| + | descriptor_fps_score_foot_comp_all_residue yes | ||
| + | descriptor_fps_score_receptor_filename ../001.files/4f4p_rec_h.mol2 | ||
| + | descriptor_fps_score_vdw_att_exp 6 | ||
| + | descriptor_fps_score_vdw_rep_exp 12 | ||
| + | descriptor_fps_score_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_score_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_score_dielectric 4.0 | ||
| + | descriptor_fps_score_vdw_fp_scale 1 | ||
| + | descriptor_fps_score_es_fp_scale 1 | ||
| + | descriptor_fps_score_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename ../004.dock/4f4p.lig.min_scored.mol2 | ||
| + | descriptor_hms_score_ref_filename ../004.dock/4f4p.lig.min_scored.mol2 | ||
| + | descriptor_hms_score_matching_coeff -5 | ||
| + | descriptor_hms_score_rmsd_coeff 1 | ||
| + | descriptor_volume_score_reference_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 | ||
| + | descriptor_volume_score_overlap_compute_method analytical | ||
| + | descriptor_weight_cont_score 1 | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
| + | descriptor_weight_fingerprint_tanimoto -1 | ||
| + | descriptor_weight_hms_score 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn | ||
| + | pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | Once you done the rescoring correctly, you will get several output files as below. | ||
| + | descriptor.output_footprint_scored.txt | ||
| + | descriptor.output_scored.mol2 | ||
| + | descriptor.output_hbond_scored.txt | ||
| + | rescore.out | ||
| + | |||
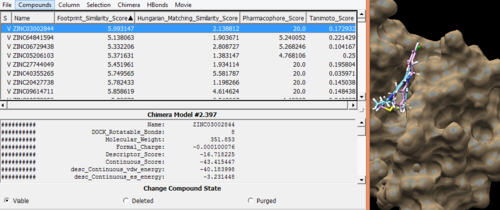
| + | Open up the descriptor.output_scored.mol2 file along with original ligand and receptor and analyse the best ligands based on different scoring functions using viewdock property in Chimera. | ||
| + | |||
| + | [[File:Rescore.png|thumb|center|500px|Figure 16: Rescored ligands out file along with Receptor and original ligand (sorted according to the footprint similarity score values)]] | ||
Latest revision as of 13:10, 9 March 2020
This tutorial teaches you how to dock a drug molecule to a receptor.
/*This page is under construction*/
Contents
I. Introduction
DOCK
DOCK is an important molecular docking program in molecular modeling and drug design. It enables you to dock a small molecule (ligand) to a binding pocket of certain protein target (receptor) and find out correct binding geometry, types and number of interactions involve in binding and energies associate with binding. For this tutorial DOCK6.9 package was used. Purpose of using DOCK6.9 in this tutorial is to re-dock a ligand to a binding pocket of the receptor protein for pose reproduction and find out lead compounds which have stronger binding affinity with our receptor binding site.
Virtual Screening
This is a protocol widely used in drug design which enable you to find out small molecules that have higher binding affinity with the binding pocket of protein of interest by screening a millions of compounds in a virtual database (such as ZINC).
Organization of directories
Log in to the seawulf cluster and go to your group directory. Use following command to create files.
mkdir 001.files 002.surface_spheres 003.gridbox 004.dock 005.virtual_screen 006.virtual_screen_mpi 007.cartesianmin 008.rescore
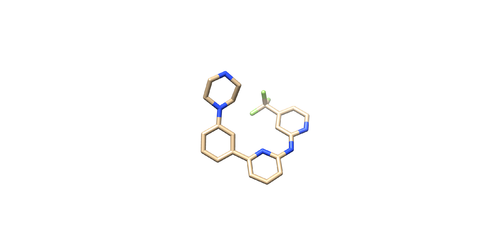
4F4P
4F4P is the crystal structure of Tyrosine Kinase SYK in complex with ligand LASW836. You can get the pdb file from here [1]. The resolution is 2.37 Å. Click Download Files -> PDB Format to download PDB file.
II. Preparation of the ligand and receptor
Checking the structure
Read the article related to the PDB file [2] to understand protonation states, charges and other important information regarding the receptor and the ligand.
Open the pdb file through chimera and look at the structure.
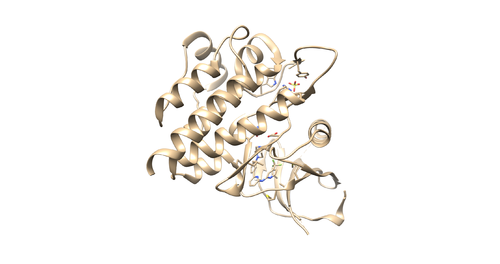
Identify the main components of the model (receptor, ligand, solvent, cofactors). In our case, the PDB file contains one protein chain, ligand(OSB), sulfate ion(SO4(2-)) as a cofactor and water molecules (H2O).
Next, we are going to generate the receptor file and the ligand file.
Prepare the Receptor file
With the PDB file loaded by chimera, do
Select -> Residue -> SO4
to select the sulfate. Use
Actions -> Atoms/Bonds -> Delete
to delete it. Then do
Select -> Residue -> HOH
to select the water molecules. Use the same method to delete it. Then do
Select -> Structure -> (0SB)
to select ligand molecule. Use the same method to delete it. Save the receptor as 4f4p_rec_noh.mol2.
OR
With the PDB file loaded by chimera, hold the control button and click on the protein chain to select a protein residue. Then click up arrow button to select the whole protein chain (Receptor). Then
Select -> Invert(all models)
to invert your selection (This will select all the components in your PDB file except receptor). Then do
Actions->Atoms/bonds->Delete.
You will remain with a protein receptor. Save the receptor as 4f4p_rec_noh.mol2.
Prepare the Ligand File
Open the pdf file again. Do
Select -> Structure -> ligand(0SB)
to select the ligand. Do
Select -> Invert (all models),
then delete the selected atoms. This will left the ligand molecule only. Save it as 4f4p_lig_noh.mol2.
Adding hydrogen and charge
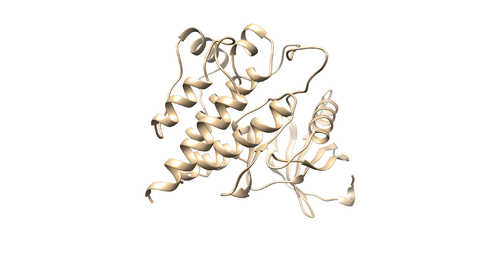
Now we are going to add hydrogen atoms and charges to our receptor and ligand. Open 4f4p_rec_noh.mol2 file using Chimera and use the following instructions to prepare the receptor file to be used in DOCK. Tools -> Structure editing -> AddH. This command will add hydrogen to the receptor. Tools -> Structure editing -> Add Charge. This command will add charge to atoms to make the receptor neutral. Save the file as 4f4p_rec_h.mol2
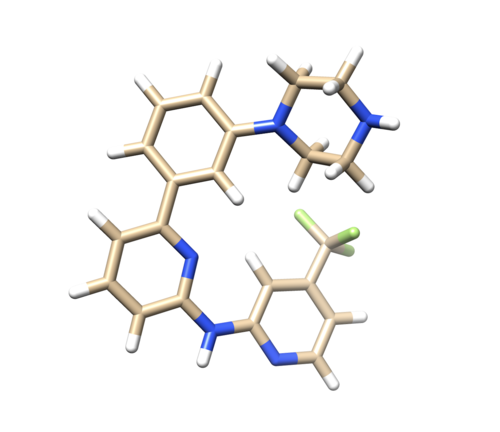
Open 4f4p_lig_noh.mol2, follow the same steps for ligand file, add hydrogen. The Chimera added 2 hydrogens to the nitrogen where it is supposed to be only one, as referred in the pdb report. So remove one hydrogen.
(Hold Ctrl and select extra H, then Actions -> Atoms/Bonds -> delete.) (Therefore, Be really concern when adding Hydrogens using chimera since it is automatically adding unwanted hydrogen atoms. Compare your protonation state with PDB report and make sure you have correct structure after adding hydrogens.)
After removing hydrogen add charges to the ligand
Tools -> Structure editing -> Add Charge. After this save the file as 4f4p_lig_h.mol2.
copy the 4f4p_rec_h.mol2, 4f4p_rec_noh.mol2, 4f4p_lig_noh.mol2, 4f4p_lig_h.mol2 files in to 001.files directory
III. Generating receptor surface and spheres
Creating a surface (DMS) file
Open the 4f4p_rec_noh.mol2 file in chimera. Then Action -> surface -> show -> Shows the surface of the receptor without H.
Tools -> Structure editing -> write DMS -> name: 4f4p_rec_noh.dms -> This command saves the surface of the receptor in dms format.
copy the 4f4p_rec.dms file in to 002.surface_spheres directory.
Generating spheres
Go to 002.surface_spheres folder Create a new input file to create spheres by
vi INSPH
then type the following lines inside the file.
4f4p_rec_noh.dms R X 0.0 4.0 1.4 4f4p_rec.sph
The first line 4f4p_rec_noh.dms specifies the input file. R indicates that spheres generated will be outside of the receptor surface. X specifies all the points will be used. 0.0 is the distance in angstroms and it will avoid steric clashes. 4.0 is the maximum surface radius of the spheres and 1.4 is the minimum radius in angstroms.The last line 4f4p_rec.sph creates the sph file that contains clustered spheres.
Once the INSPH file is ready, type the following command to generate the spheres.
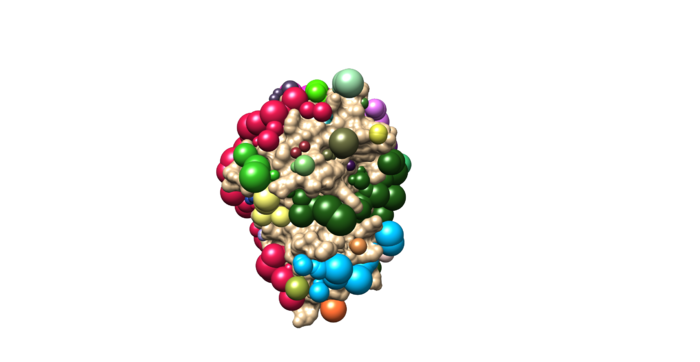
sphgen -i INSPH -o OUTSPH
Once sphgen command is successful, following output files will be created.
OUTSPH 4f4p_rec.sph
Open the f4p_rec.sph using Chimera along with 4f4p_rec_noh.mol2 file. You should get a similar output like the image below.
Selecting Spheres
Here we will be selecting the spheres which defines the binding pocket of the ligand because we are trying to direct the ligand towards that binding site rather than all over the receptor. To select the spheres type the following command.
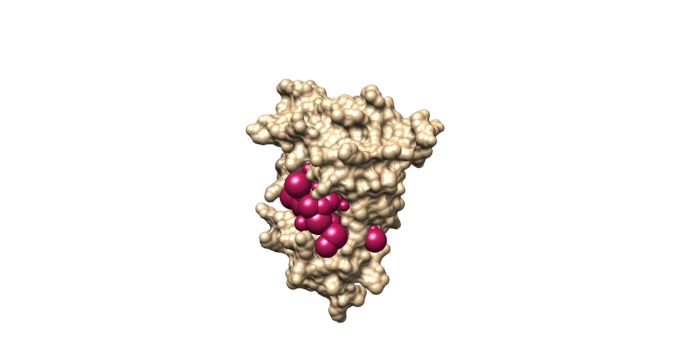
sphere_selector 4f4p_rec.sph ../001.files/4f4p_noh_lig.mol2 10.0
This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph.
Output files:
selected_spheres.sph
Visualize the selected spheres using Chimera to make sure the correct spheres are selected. Notice that, spheres around the ligand binding site are kept and all the other spheres are deleted by following sphere selector command .
IV. Generating box and grid
Generating box
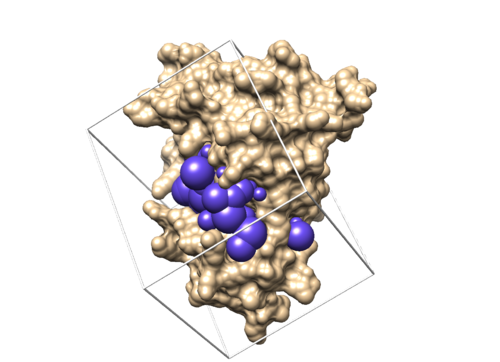
Move to 003.gridbox directory Create a new file showbox.in and write the following lines in the file(# means comment ).
Y #Say 'Yes' to make a box 8.0 #The boundary of the box is at least 8 Angstroms from any spheres ../002.surface_spheres/selected_spheres.sph #The location and file name of the sphere file 1 #Use the first cluster of spheres (only 1 cluster in this case) 4f4p.box.pdb #The output filename
Use the following command to generate the box.
showbox < showbox.in
If this step is successful, you should see a new file (4f4p.box.pdb) in 003.gridbox folder.
Output file:
4f4p.box.pdb
Generating Grid
Create a new input file called grid.in and write the following information there.
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../001.files/4f4p_rec_h.mol2 box_file 4f4p.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
save the file and run the following command.
grid -i grid.in -o grid.out
If this is successful, you will get following output files.
grid.out grid.nrg grid.bmp
V. Docking
Energy minimization
Before doing docking pose reproduction, energy minimization can be done in order to remove unfavorable intra-molecular clashes of the ligand. (If not it will affect the later steps of docking)
Go to 004.dock folder and open a file called min.in and type the following content there.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/4f4p_lig_h.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol ../001.files/4f4p_lig_h.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 4f4p.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
Save the file and run the following command.
dock6 -i min.in -o min.out
If you have done correctly, you will get followig out files.
min.out 4f4p.lig.min_scored.mol2
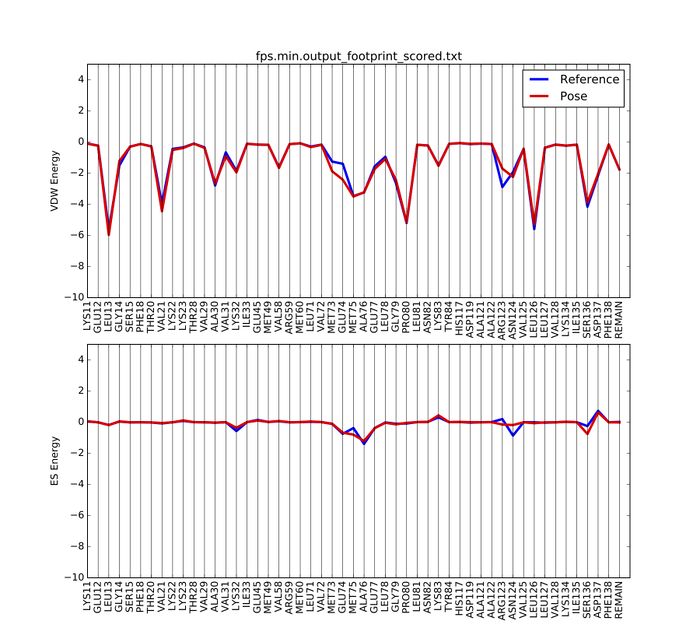
Footprint Analysis
In order to see the minimized ligand compared to the original ligand through footprint analysis, open a file called footprint.in and type following.
conformer_search_type rigid use_internal_energy no ligand_atom_file 4f4p.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ../001.files/4f4p_lig_h.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ../001.files/4f4p_rec_h.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 9 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix fps.min.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
If footprint runs successfully, you can find the following files generated
footprint.out fps.min.output_footprint_scored.txt fps.min.output_hbond_scored.txt fps.min.output_scored.mol2
In order to visualize the footprint, first we need to copy a python script into our directory, then run it:
cp /gpfs/projects/AMS536/zzz.programs/plot_footprint_single_magnitude.py ./ python plot_footprint_single_magnitude.py fps.min.output_footprint_scored.txt 50
A pdf document named fps.min.output_footprint_scored.txt.pdf will be generated. It looks like this:
Rigid Docking
Create an input file for rigid docking
touch rigid.in
Write lines below into the rigid.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 9 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/4f4p_lig_h.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/4f4p_lig_h.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 2 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 40 write_conformations yes cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Run dock using the created input file.
dock6 -i rigid.in -o rigid.out
Once rigid docking is successful, you will get following output files.
rigid.out rigid.out_conformers.mol2 rigid.out_scored.mol2
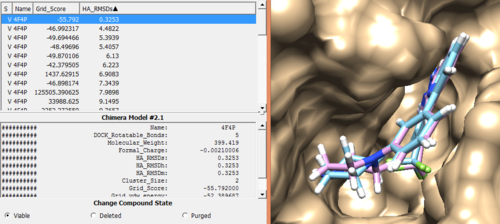
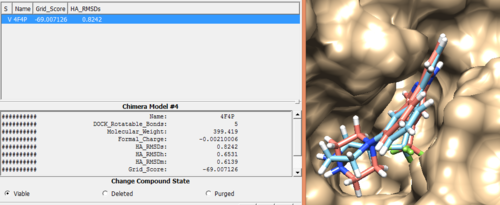
In order to check the rigid docking success, open rigid.out_scored.mol2 output file using Chimera by following steps.
Open Chimera File -> Open -> 4f4p_rec_h.mol2 (Actions -> Surface -> Show) File -> Open -> 4f4p_lig_h.mol2 Tools -> Surface/binding Analysis -> ViewDock -> Select the Rigid Dock output file. (rigid.out_scored.mol2) In the loaded dialog box select Dock4,5 or 6
Once everything is loaded go to the ViewDock window and use it's menu to view all the calculated properties regarding the rigid docked ligand by following the steps below.
Column -> Show -> Grid_Score Column -> Show -> HA_RMSDs
Follow the same steps to get all different properties as you interested.
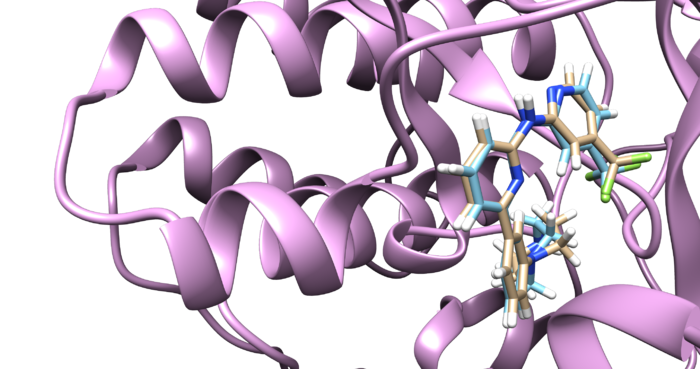
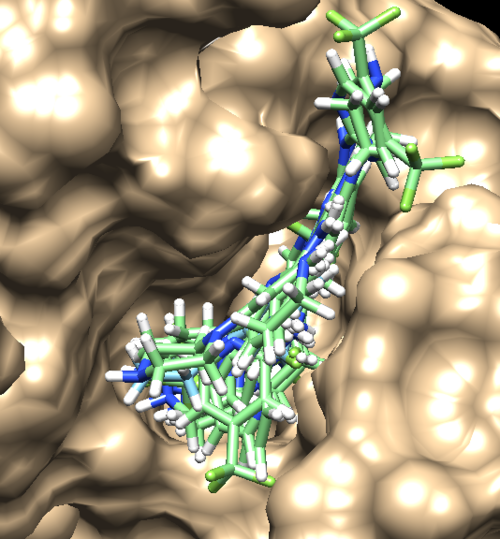
You can choose different conformations from the ViewDock window. If you sorted it, the top one should be the conformation with lowest energy. Your visualized structure should be similar to the image below.
Fixed Anchor Docking
Create an input file called fad.in and use the following command.
dock6 -i fad.in -o fad.out
Use the following lines to answer the prompted questions as we did in rigid docking.
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 9 internal_energy_cutoff 100.0 ligand_atom_file ../001.files/4f4p_lig_h.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.files/4f4p_lig_h.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 4f4p_fad write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Once docking is completed following output files will be generated.
fad.out 4f4p_fad_scored.mol2
Follow the same method used in rigid docking to visualize the docked poses using Chimera by opening the 4f4p_fad_scored.mol2 file. Once it is visualized, it should like the image below.
Flexible Docking
Create a new input file for flexible docking called flex.in and perform flexible docking using DOCK6 by following command.
dock6 -i flex.in -o flex.out
Answer the prompted questions using the following lines as we did in rigid and fixed anchor docking (or copy the following content in to flex.in file and save).
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no write_fragment_libraries no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.dock/4f4p.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.dock/4f4p.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 1 rank_ligands no
Once flexible docking is completed following output files will be generated.
flex.out flex.out_scored.mol2
Use the visualization steps used in rigid and fixed anchor docking and study the properties of the docking results by opening flex.out_scored.mol2 file. (You will get one confirmation since you're writing num_scored_conformers as one in the above script.)
VI. Virtual Screen
Virtual screening is the method of screening a ligand library (drug-like molecules) to filter the best ligands which can bind to the binding site of a specific receptor. Here we will be using a ligand library which contains 25000 molecules to select the best ligands which can be used to replace the original ligand that the PDB file contained.
Move to the directory 005.virtual_screen.
Copy the ligand library to the same directory.
cp /gpfs/projects/AMS536/2020/536_class/VS_library_5K.mol2 ./
Create a new input file for virtual screen.
touch virtual.in
Use the input file to perform virtual screen using DOCK6.
dock6 -i virtual.in -o virtual.out
Use the following lines to answer the prompted questions.
conformer_search_type flex
write_fragment_libraries no
user_specified_anchor no
limit_max_anchors no
min_anchor_size 5
pruning_use_clustering yes
pruning_max_orients 1000
pruning_clustering_cutoff 100
pruning_conformer_score_cutoff 100.0
pruning_conformer_score_scaling_factor 1.0
use_clash_overlap no
write_growth_tree no
use_internal_energy yes
internal_energy_rep_exp 9
internal_energy_cutoff 100.0
ligand_atom_file VS_library_5K.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd no
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file ../002.surface_spheres/selected_spheres.sph
max_orientations 1000
critical_points no
chemical_matching no
use_ligand_spheres no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../003.gridbox/grid
multigrid_score_secondary no
dock3.5_score_secondary no
continuous_score_secondary no
footprint_similarity_score_secondary no
pharmacophore_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_score_secondary no
amber_score_secondary no
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_anchor_max_iterations 500
simplex_grow_max_iterations 500
simplex_grow_tors_premin_iterations 0
simplex_random_seed 0
simplex_restraint_min no
atom_model all
vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn
flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn
flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl
ligand_outfile_prefix virtual.out
write_orientations no
num_scored_conformers 1
rank_ligands no
You can run this job on head node for a short while, just to make sure everything is fine. Then you can kill the job by Ctrl-C.
VII Virtual Screeni mpi
Go to 006. virtual_screen_mpi directory and copy the virtual.in file here. We will run the job on the seawulf cluster using slurm script.
Creat the virtual.sh file
vi virtual.sh
Add the following lines there
#!/bin/sh #SBATCH --partition=long-40core #SBATCH --time=48:00:00 #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --job-name=........#whatever you want #SBATCH --output=%x-%j.o dock6 -i virtual.in -o virtual.out
Submit the job on seawulf using
sbatch virtual.sh
To check the status of your job
squeue -u [NETID]
Check Virtual Screen output
The job was terminated due to time limit. In order to calculate how many molecules were screened, we run the command:
grep "Molecule: ZINC" virtual.out | wc -l
Everyone in our group has got either 1066, 1068, 1061 molecules processed.
To vizualize the output, we open the file virtual.out_scored.mol2 with Chimera. The Chimera could be slow as there are many many molecules. Go to Tools -> Surface binding Analysis -> View Dock. Also open the receptor in this session.
Column -> Show -> Grid Score
Column -> Show -> Mass weight
We see that best Grid scored molecules have higher energy than those that have lower Grid score energy. The vdw energy also have the same trend with Grid score.
VIII.Cartesian Minimization
Here we will use the docked molecules and perform a cartesian minimization of them.
Move to the directory 007.cartesianmin.
Create a new input file for the minimization.
touch min.in
Use the input file to perform the cartesian minimization using DOCK6.
dock6 -i min.in -o min.out
Use the following lines to answer the prompted questions.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../005.virtual_screen/virtual.out_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename ../001.files/4f4p_rec_h.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 4f4p.virtualscreen.minimized write_orientations no num_scored_conformers 1 rank_ligands no
Then in order to submit the script in to seawulf cluster, open a new script called cartesianmin.sh and write the following.
#!/bin/sh #SBATCH --partition=long-40core #SBATCH --time=48:00:00 #SBATCH --nodes=1 #SBATCH --ntasks=1 #SBATCH --job-name=4f4p_cartesian #SBATCH --output=%x-%j.o
echo "============================= SLURM JOB =================================" date echo echo " The job will be started on the following node(s):" echo $SLURM_JOB_NODELIST echo echo "Slurm user: $SLURM_JOB_USER" echo "Run directory: $(pwd)" echo "Job ID: $SLURM_JOB_ID" echo "Job name: $SLURM_JOB_NAME" echo "Partition: $SLURM_JOB_PARTITION" echo "Number of nodes: $SLURM_JOB_NUM_NODES" echo "Number of tasks: $SLURM_NTASKS" echo "Submitted from: $SLURM_SUBMIT_HOST:$SLURM_SUBMIT_DIR" echo "========================================================================="
dock6 -i min.in -o min.out
Then submit the job into seawulf cluster.
sbatch cartesianmin.sh
Once it has done correctly, you will get a output file called 4f4p.virtualscreen.minimized_scored.mol2. This file contains molecules with minimized energies by removing unfavorable steric clashes.
IX Rescoring Docked Molecules
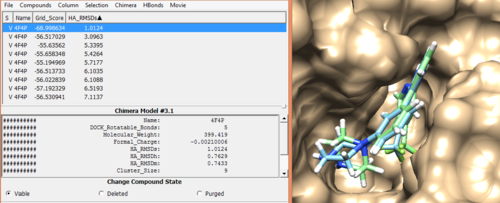
In order to rank our docked ligands and extract the best ligands (which has the most negative, lowest scores) based on different scoring functions like Footprint similarity, pharmacophore score, tanimoto score, the hungarian and the volume overlap score, we will be using Dock6 to rescore the virtual screen minimized ligands.
Move to the directory 008.rescore and create a new input file for the rescoring.
touch rescore.in
Use the input file to perform the cartesian minimization using DOCK6.
dock6 -i rescore.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../007.cartesianmin/4f4p.virtualscreen.minimized_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score yes descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_cont_score_rec_filename ../001.files/4f4p_rec_h.mol2 descriptor_cont_score_att_exp 6 descriptor_cont_score_rep_exp 12 descriptor_cont_score_rep_rad_scale 1 descriptor_cont_score_use_dist_dep_dielectric yes descriptor_cont_score_dielectric 4.0 descriptor_cont_score_vdw_scale 1 descriptor_cont_score_es_scale 1 descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../001.files/4f4p_rec_h.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../004.dock/4f4p.lig.min_scored.mol2 descriptor_hms_score_ref_filename ../004.dock/4f4p.lig.min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename ../004.dock/4f4p.lig.min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_cont_score 1 descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Once you done the rescoring correctly, you will get several output files as below.
descriptor.output_footprint_scored.txt descriptor.output_scored.mol2 descriptor.output_hbond_scored.txt rescore.out
Open up the descriptor.output_scored.mol2 file along with original ligand and receptor and analyse the best ligands based on different scoring functions using viewdock property in Chimera.