Difference between revisions of "2021 Denovo tutorial 3 with PDBID 1S19"
Stonybrook (talk | contribs) (→Fragment Libraries) |
Stonybrook (talk | contribs) (→Generic Denovo Design with PDB 1S19) |
||
| (16 intermediate revisions by the same user not shown) | |||
| Line 30: | Line 30: | ||
orient_ligand yes | orient_ligand yes | ||
automated_matching yes | automated_matching yes | ||
| − | receptor_site_file ../ | + | receptor_site_file ../002.surface_spheres/selected_spheres.sph |
max_orientations 1000 | max_orientations 1000 | ||
critical_points no | critical_points no | ||
| Line 52: | Line 52: | ||
Once this has ran, the following files should have been generated: | Once this has ran, the following files should have been generated: | ||
| − | '''fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat''' | + | '''fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat'''. To check the number of fragments in each file, you can type: |
| − | To check the number of fragments in each file, you can type: | ||
grep -wc MOLECULE *.mol2 | grep -wc MOLECULE *.mol2 | ||
| + | |||
| + | You should get the following output upon using this command: | ||
| + | |||
| + | fraglib_linker.mol2:4 | ||
| + | fraglib_rigid.mol2:0 | ||
| + | fraglib_scaffold.mol2:2 | ||
| + | fraglib_sidechain.mol2:2 | ||
| + | fragment.out_scored.mol2:0 | ||
| + | |||
| + | |||
| + | These fragments can also be visualized in Chimera. The results are shown below. | ||
| + | |||
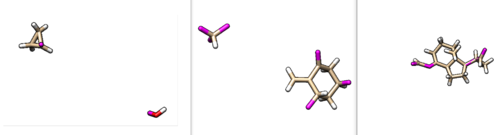
| + | [[File:frag_three.png|thumb|center|500px|2p16 fragments. The left image is the side chain fragments, the center are the scaffold fragments and the right image is the linker fragment.]] | ||
===Focused Denovo Growth=== | ===Focused Denovo Growth=== | ||
| + | |||
| + | We will be using the fragments that we have generated for the denovo dock. The different fragments will be connected together using restraints such as molecular weight and formal charges. DOCK will check the torsion environment file that was created in the fragment generation section to ensure reasonable connections are being made. | ||
| + | |||
| + | To begin, we will make a new directory and call it denovo_focus | ||
| + | |||
| + | mkdir dn_focus | ||
| + | |||
| + | Create a file called '''focus.in''' and copy the following script into the file. | ||
| + | |||
| + | conformer_search_type denovo | ||
| + | dn_fraglib_scaffold_file ../fraglib/fraglib_scaffold.mol2 | ||
| + | dn_fraglib_linker_file ../fraglib/fraglib_linker.mol2 | ||
| + | dn_fraglib_sidechain_file ../fraglib/fraglib_sidechain.mol2 | ||
| + | dn_user_specified_anchor no | ||
| + | dn_use_torenv_table yes | ||
| + | dn_torenv_table ../fraglib/fraglib_torenv.dat | ||
| + | dn_sampling_method graph | ||
| + | dn_graph_max_picks 30 | ||
| + | dn_graph_breadth 3 | ||
| + | dn_graph_depth 2 | ||
| + | dn_graph_temperature 100.0 | ||
| + | dn_pruning_conformer_score_cutoff 100.0 | ||
| + | dn_pruning_conformer_score_scaling_factor 1.0 | ||
| + | dn_pruning_clustering_cutoff 100.0 | ||
| + | dn_constraint_mol_wt 550.0 | ||
| + | dn_constraint_rot_bon 15 | ||
| + | dn_constraint_formal_charge 2.0 | ||
| + | dn_heur_unmatched_num 1 | ||
| + | dn_heur_matched_rmsd 2.0 | ||
| + | dn_unique_anchors 1 | ||
| + | dn_max_grow_layers 9 | ||
| + | dn_max_root_size 25 | ||
| + | dn_max_layer_size 25 | ||
| + | dn_max_current_aps 5 | ||
| + | dn_max_scaffolds_per_layer 1 | ||
| + | dn_write_checkpoints yes | ||
| + | dn_write_prune_dump no | ||
| + | dn_write_orients no | ||
| + | dn_write_growth_trees no | ||
| + | dn_output_prefix dn_focus.out | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../002.surface_spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../003.gridbox/grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file | ||
| + | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
===Focused Denovo Rescore=== | ===Focused Denovo Rescore=== | ||
| + | |||
| + | Now that all of the ligands have been generated in silico, we will want to rescore them based on their binding properties to predict which ones would be the best binders in vitro. | ||
| + | |||
| + | First, create a new directory and dock6 input file: | ||
| + | |||
| + | mkdir 12.dn_focus_rescore | ||
| + | touch dn_rescore.in | ||
| + | |||
| + | Define parameters in the dn_rescore.in file as follows and submit the job: | ||
| + | |||
| + | conformer_search_type rigid | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../dn_focus/dn_focus.out.denovo_build.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score no | ||
| + | descriptor_use_multigrid_score no | ||
| + | descriptor_use_continuous_score no | ||
| + | descriptor_use_footprint_similarity yes | ||
| + | descriptor_use_pharmacophore_score yes | ||
| + | descriptor_use_tanimoto yes | ||
| + | descriptor_use_hungarian yes | ||
| + | descriptor_use_volume_overlap yes | ||
| + | descriptor_fps_score_use_footprint_reference_mol2 yes | ||
| + | descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 | ||
| + | descriptor_fps_score_foot_compare_type Euclidean | ||
| + | descriptor_fps_score_normalize_foot no | ||
| + | descriptor_fps_score_foot_comp_all_residue yes | ||
| + | descriptor_fps_score_receptor_filename ../001.structure/1s19.dockprep.mol2 | ||
| + | descriptor_fps_score_vdw_att_exp 6 | ||
| + | descriptor_fps_score_vdw_rep_exp 12 | ||
| + | descriptor_fps_score_vdw_rep_rad_scale 1 | ||
| + | descriptor_fps_score_use_distance_dependent_dielectric yes | ||
| + | descriptor_fps_score_dielectric 4.0 | ||
| + | descriptor_fps_score_vdw_fp_scale 1 | ||
| + | descriptor_fps_score_es_fp_scale 1 | ||
| + | descriptor_fps_score_hb_fp_scale 0 | ||
| + | descriptor_fms_score_use_ref_mol2 yes | ||
| + | descriptor_fms_score_ref_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 | ||
| + | descriptor_fms_score_write_reference_pharmacophore_mol2 no | ||
| + | descriptor_fms_score_write_reference_pharmacophore_txt no | ||
| + | descriptor_fms_score_write_candidate_pharmacophore no | ||
| + | descriptor_fms_score_write_matched_pharmacophore no | ||
| + | descriptor_fms_score_compare_type overlap | ||
| + | descriptor_fms_score_full_match yes | ||
| + | descriptor_fms_score_match_rate_weight 5.0 | ||
| + | descriptor_fms_score_match_dist_cutoff 1.0 | ||
| + | descriptor_fms_score_match_proj_cutoff 0.7071 | ||
| + | descriptor_fms_score_max_score 20 | ||
| + | descriptor_fingerprint_ref_filename ../004.dock/1s19_lig.min_scored.mol2 | ||
| + | descriptor_hms_score_ref_filename ../004.dock/1s19_lig.min_scored.mol2 | ||
| + | descriptor_hms_score_matching_coeff -5 | ||
| + | descriptor_hms_score_rmsd_coeff 1 | ||
| + | descriptor_volume_score_reference_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 | ||
| + | descriptor_volume_score_overlap_compute_method analytical | ||
| + | descriptor_weight_fps_score 1 | ||
| + | descriptor_weight_pharmacophore_score 1 | ||
| + | descriptor_weight_fingerprint_tanimoto -1 | ||
| + | descriptor_weight_hms_score 1 | ||
| + | descriptor_weight_volume_overlap_score -1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand no | ||
| + | atom_model all | ||
| + | vdw_defn_file | ||
| + | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn | ||
| + | pharmacophore_defn_file | ||
| + | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn | ||
| + | ligand_outfile_prefix descriptor.output | ||
| + | write_footprints yes | ||
| + | write_hbonds yes | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | dock6 -i dn_rescore.in -o dn_rescore.out | ||
| + | |||
| + | It may be helpful to submit this job using a slurm script. An example is shown below: | ||
| + | |||
| + | #!/bin/bash | ||
| + | #SBATCH --time=48:00:00 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --ntasks=28 | ||
| + | #SBATCH --job-name=dn_rescore | ||
| + | #SBATCH --output=dn_rescore.out | ||
| + | #SBATCH -p long-28core | ||
| + | |||
| + | cd $SLURM_SUBMIT_DIR | ||
| + | echo "starting Dock6.9 simulation" | ||
| + | /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i dn_rescore.in -o dn_rescore.out | ||
= Generic Denovo Design with PDB 1S19 = | = Generic Denovo Design with PDB 1S19 = | ||
| + | |||
| + | With focused docking, we only used fragments from the original ligand. With generic docking, we instead using a library of fragments and linkers to make new ligands. We will use the following input: | ||
| + | |||
| + | mkdir 13_dn_generic_denovo | ||
| + | touch gn_denovo.in | ||
| + | dock6 -i gn_denovo.in | ||
| + | |||
| + | conformer_search_type denovo | ||
| + | dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_scaffold.mol2 | ||
| + | dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_linker.mol2 | ||
| + | dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_sidechain.mol2 | ||
| + | dn_user_specified_anchor no | ||
| + | dn_use_torenv_table yes | ||
| + | dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_torenv.dat | ||
| + | dn_sampling_method graph | ||
| + | dn_graph_max_picks 30 | ||
| + | dn_graph_breadth 3 | ||
| + | dn_graph_depth 2 | ||
| + | dn_graph_temperature 100 | ||
| + | dn_pruning_conformer_score_cutoff 100 | ||
| + | dn_pruning_conformer_score_scaling_factor 1.0 | ||
| + | dn_pruning_clustering_cutoff 100.0 | ||
| + | dn_constraint_mol_wt 550.0 | ||
| + | dn_constraint_rot_bon 15 | ||
| + | dn_constraint_formal_charge 2.0 | ||
| + | dn_heur_unmatched_num 1 | ||
| + | dn_heur_matched_rmsd 2.0 | ||
| + | dn_unique_anchors 1 | ||
| + | dn_max_grow_layers 9 | ||
| + | dn_max_root_size 25 | ||
| + | dn_max_layer_size 25 | ||
| + | dn_max_current_aps 5 | ||
| + | dn_max_scaffolds_per_layer 1 | ||
| + | dn_write_checkpoints yes | ||
| + | dn_write_prune_dump no | ||
| + | dn_write_orients no | ||
| + | dn_write_growth_trees no | ||
| + | dn_output_prefix gn_denovo_output | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../002.surface_spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | multigrid_score_primary no | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary no | ||
| + | continuous_score_secondary no | ||
| + | footprint_similarity_score_primary no | ||
| + | footprint_similarity_score_secondary no | ||
| + | pharmacophore_score_primary no | ||
| + | pharmacophore_score_secondary no | ||
| + | descriptor_score_primary yes | ||
| + | descriptor_score_secondary no | ||
| + | descriptor_use_grid_score yes | ||
| + | descriptor_use_pharmacophore_score no | ||
| + | descriptor_use_tanimoto no | ||
| + | descriptor_use_hungarian no | ||
| + | descriptor_use_volume_overlap no | ||
| + | descriptor_grid_score_rep_rad_scale 1 | ||
| + | descriptor_grid_score_vdw_scale 1 | ||
| + | descriptor_grid_score_es_scale 1 | ||
| + | descriptor_grid_score_grid_prefix ../003.gridbox/grid | ||
| + | descriptor_weight_grid_score 1 | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl | ||
| + | |||
| + | If we look at our output file gn_denovo.out, we see that 60285 molecules were processed in 2478 seconds using SLURM. You can visualize these molecules and their corresponding binding interactions using ViewDock in Chimera as before. | ||
Latest revision as of 16:24, 15 March 2021
This tutorial continues where we left off from the virtual screening of 1S19, a structure of the vitamin D nuclear receptor bound to the side chain analogue calcipotriol.
Contents
Focused Denovo Design with PDB 1S19
In this section, we will generate new ligand structures using a fragment library. It will require the use of some of the output files from the Virtual Screen tutorial (found here).
Fragment Libraries
We will use a focused fragment library file based on the original ligand. To begin, we will create a new directory for this.
mkdir fraglib
Create a new file called fragment.in with the following inputs.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001_structure/1s19_ligand_dockprep.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix fragment.out write_orientations no num_scored_conformers 1 rank_ligands no
To run this, type:
dock6 -i fragment.in -o fragment.out
Once this has ran, the following files should have been generated: fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat. To check the number of fragments in each file, you can type:
grep -wc MOLECULE *.mol2
You should get the following output upon using this command:
fraglib_linker.mol2:4 fraglib_rigid.mol2:0 fraglib_scaffold.mol2:2 fraglib_sidechain.mol2:2 fragment.out_scored.mol2:0
These fragments can also be visualized in Chimera. The results are shown below.
Focused Denovo Growth
We will be using the fragments that we have generated for the denovo dock. The different fragments will be connected together using restraints such as molecular weight and formal charges. DOCK will check the torsion environment file that was created in the fragment generation section to ensure reasonable connections are being made.
To begin, we will make a new directory and call it denovo_focus
mkdir dn_focus
Create a file called focus.in and copy the following script into the file.
conformer_search_type denovo dn_fraglib_scaffold_file ../fraglib/fraglib_scaffold.mol2 dn_fraglib_linker_file ../fraglib/fraglib_linker.mol2 dn_fraglib_sidechain_file ../fraglib/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table ../fraglib/fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix dn_focus.out use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl
Focused Denovo Rescore
Now that all of the ligands have been generated in silico, we will want to rescore them based on their binding properties to predict which ones would be the best binders in vitro.
First, create a new directory and dock6 input file:
mkdir 12.dn_focus_rescore touch dn_rescore.in
Define parameters in the dn_rescore.in file as follows and submit the job:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../dn_focus/dn_focus.out.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../001.structure/1s19.dockprep.mol2 descriptor_fps_score_vdw_att_exp 6 descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../004.dock/1s19_lig.min_scored.mol2 descriptor_hms_score_ref_filename ../004.dock/1s19_lig.min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename ../004.dock/1s19.lig.min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/ph4.defn ligand_outfile_prefix descriptor.output write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
dock6 -i dn_rescore.in -o dn_rescore.out
It may be helpful to submit this job using a slurm script. An example is shown below:
#!/bin/bash #SBATCH --time=48:00:00 #SBATCH --nodes=1 #SBATCH --ntasks=28 #SBATCH --job-name=dn_rescore #SBATCH --output=dn_rescore.out #SBATCH -p long-28core cd $SLURM_SUBMIT_DIR echo "starting Dock6.9 simulation" /gpfs/projects/AMS536/zzz.programs/dock6.9_release/bin/dock6.mpi -i dn_rescore.in -o dn_rescore.out
Generic Denovo Design with PDB 1S19
With focused docking, we only used fragments from the original ligand. With generic docking, we instead using a library of fragments and linkers to make new ligands. We will use the following input:
mkdir 13_dn_generic_denovo touch gn_denovo.in dock6 -i gn_denovo.in
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100 dn_pruning_conformer_score_cutoff 100 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix gn_denovo_output use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score yes descriptor_use_pharmacophore_score no descriptor_use_tanimoto no descriptor_use_hungarian no descriptor_use_volume_overlap no descriptor_grid_score_rep_rad_scale 1 descriptor_grid_score_vdw_scale 1 descriptor_grid_score_es_scale 1 descriptor_grid_score_grid_prefix ../003.gridbox/grid descriptor_weight_grid_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl
If we look at our output file gn_denovo.out, we see that 60285 molecules were processed in 2478 seconds using SLURM. You can visualize these molecules and their corresponding binding interactions using ViewDock in Chimera as before.