Difference between revisions of "Pose Reproduction SB2024 V1 DOCK6.10 A"
(→II.Necessary files) |
m (Ccorbo moved page Pose Reproduction to Pose Reproduction SB2024 V1 DOCK6.10 A) |
||
| (31 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
The purpose of this tutorial is to develop a uniform method to test pose reproduction across the Rizzo lab with the DOCK software. Note any data in this tutorial is solely for the purpose of example. | The purpose of this tutorial is to develop a uniform method to test pose reproduction across the Rizzo lab with the DOCK software. Note any data in this tutorial is solely for the purpose of example. | ||
| Line 17: | Line 16: | ||
==II.Necessary files== | ==II.Necessary files== | ||
| − | This tutorial uses Single Grid Energy as the primary score for docking | + | Scripts to run Pose Reproduction in batch mode are found at: |
| + | https://github.com/rizzolab/Benchmarking_and_Validation | ||
| + | |||
| + | This tutorial uses Single Grid Energy as the primary score for docking. Thus, grid files are required. The receptor mol2 is only necessary for visualization purposes. | ||
The list of necessary files are: | The list of necessary files are: | ||
${pdb_id}.lig.am1bcc.mol2 | ${pdb_id}.lig.am1bcc.mol2 | ||
| Line 25: | Line 27: | ||
${pdb_id}.rec.nrg | ${pdb_id}.rec.nrg | ||
| − | All necessary files are available for download at: | + | All necessary files for different versions of our test set are available for download at: |
| − | To | + | https://ringo.ams.stonybrook.edu/index.php/Rizzo_Lab_Downloads |
| + | |||
| + | To (re)create a testset using Rizzo Lab Protocols: | ||
| + | https://ringo.ams.stonybrook.edu/index.php/Test_Set_Tutorial_V1 | ||
==III.Docking molecules== | ==III.Docking molecules== | ||
| + | Enter Pose Reproduction Directory: | ||
| + | cd Benchmarking_and_Validation/PoseReproduction/ | ||
| + | |||
| + | In 001.submit_dock.sh edit the following variables: | ||
| + | |||
| + | system_file="List of PDB codes in a file delimited by line" | ||
| + | testset="Path to necessary files for docking (section above)" | ||
| + | dock_dir="Uppermost directory of DOCK6 executable" | ||
| + | |||
| + | Additionally other variables can be changed in 001.submit_dock.sh : | ||
| + | |||
| + | condition="Unique name given to each experiment output - otherwise 'Default' " | ||
| + | seed="Random seed - otherwise '0' " | ||
| + | |||
| + | This calls the script FLX.sh for each system which writes a dock input file and then immediately calls DOCK6. | ||
| + | A separate dock input file is written for each system. You can use parameters from the input file outlined in FLX.sh, but best practice would be to '''develop a new input file for FLX.sh by first interactively creating an input file with the version of DOCK being used. The current version of FLX.sh was written for DOCK6.10 .''' This will prevent any changes in queries being overlooked: | ||
| + | touch dock_interactive.in | ||
| + | dock6 -i dock_interactive.in | ||
| + | Go through interactive question tree, using same parameters (answers to prompts) in original FLX.sh, where applicable. Once dock_interactive.in is complete, make sure the testset path and pdb id match the format and same variable names as FLX.sh : | ||
| + | ligand_atom_file /${system_dir}/${system}/${system}.lig.am1bcc.mol2 | ||
| + | |||
| + | Insert dock_interactive.in into FLX.sh, replacing the input file already in FLX.sh . | ||
| + | |||
| + | Submit the job after specifying partition and wall clock criteria. Typically ~2 minutes per system per core is sufficient. | ||
| + | sbatch 001.submit_dock.sh | ||
| + | |||
| + | When docking is completed you will have a separate directory for each system. In each directory will be the input file, output file, and mol2 of docked results. If condition was "Default" and seed was "0", the file will be named: | ||
| + | ${pdb_id}/Default_0_scored.mol2 | ||
==IV.Pose Reproduction Analysis== | ==IV.Pose Reproduction Analysis== | ||
| + | Next run a script which calculates outcomes. This script is compatible with python 2. | ||
| + | module load py/2.7.15 | ||
| + | python calculate_dock6.results.py ${condition}_${seed} ${system_file} | ||
| + | |||
| + | e.g.: python calculate_dock6.results.py Default_0 clean.systems.all | ||
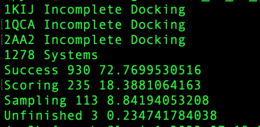
| − | + | In the below image, the systems "1KIJ","1QCA","2AA2" did not successfully dock. There were 1,279 systems in the list provided. The raw number of systems and percentage for "Success", "Score Fail" and "Sample Fail" are given. Incomplete docked systems are counted as Sample Fails. | |
| + | |||
| + | [[Image:PR_results_V1ccorbo.png|thumb|center|260px|Output from calculate_dock6.results.py shows systems which didn't dock and Success and Fail rates]] | ||
-SEE README FILE IN GIT REPO FOR ADDTIONAL DETAILS THAT MAY NOT BE COVERED HERE | -SEE README FILE IN GIT REPO FOR ADDTIONAL DETAILS THAT MAY NOT BE COVERED HERE | ||
| Line 41: | Line 81: | ||
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> | >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> | ||
| − | Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (2024) | + | Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (This tutorial was last updated 02/22/2024) |
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> | >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> | ||
Latest revision as of 16:35, 13 March 2024
The purpose of this tutorial is to develop a uniform method to test pose reproduction across the Rizzo lab with the DOCK software. Note any data in this tutorial is solely for the purpose of example.
Contents
I.Introduction
-Pose reproduction is an experiment which tests a docking programs ability to predict the bound pose of a ligand to a receptor (typically a protein). An experimental structure of a protein-ligand complex is converted into 2 separate files, 1 for ligand and 1 for receptor. The docking program then predicts the binding orientation that is most energetically favorable. In the case of DOCK6, the ligand is flexibly docked with the Anchor & Grow algorithm to a rigid receptor.
-The RMSD between the docked poses and experimental pose are measured. We consider RMSD < 2 angstroms an accurate prediction. There are 3 outcomes we classify.
1) Success - The best scoring pose is < 2 angstroms
2) Scoring Fail - A pose < 2 angstroms was sampled but did not score best
3) Sampling Fail - No pose < 2 angstroms was sampled
II.Necessary files
Scripts to run Pose Reproduction in batch mode are found at:
https://github.com/rizzolab/Benchmarking_and_Validation
This tutorial uses Single Grid Energy as the primary score for docking. Thus, grid files are required. The receptor mol2 is only necessary for visualization purposes. The list of necessary files are:
${pdb_id}.lig.am1bcc.mol2
${pdb_id}.rec.clust.close.sph
${pdb_id}.rec.clean.mol2
${pdb_id}.rec.bmp
${pdb_id}.rec.nrg
All necessary files for different versions of our test set are available for download at:
https://ringo.ams.stonybrook.edu/index.php/Rizzo_Lab_Downloads
To (re)create a testset using Rizzo Lab Protocols:
https://ringo.ams.stonybrook.edu/index.php/Test_Set_Tutorial_V1
III.Docking molecules
Enter Pose Reproduction Directory:
cd Benchmarking_and_Validation/PoseReproduction/
In 001.submit_dock.sh edit the following variables:
system_file="List of PDB codes in a file delimited by line"
testset="Path to necessary files for docking (section above)"
dock_dir="Uppermost directory of DOCK6 executable"
Additionally other variables can be changed in 001.submit_dock.sh :
condition="Unique name given to each experiment output - otherwise 'Default' "
seed="Random seed - otherwise '0' "
This calls the script FLX.sh for each system which writes a dock input file and then immediately calls DOCK6.
A separate dock input file is written for each system. You can use parameters from the input file outlined in FLX.sh, but best practice would be to develop a new input file for FLX.sh by first interactively creating an input file with the version of DOCK being used. The current version of FLX.sh was written for DOCK6.10 . This will prevent any changes in queries being overlooked:
touch dock_interactive.in
dock6 -i dock_interactive.in
Go through interactive question tree, using same parameters (answers to prompts) in original FLX.sh, where applicable. Once dock_interactive.in is complete, make sure the testset path and pdb id match the format and same variable names as FLX.sh :
ligand_atom_file /${system_dir}/${system}/${system}.lig.am1bcc.mol2
Insert dock_interactive.in into FLX.sh, replacing the input file already in FLX.sh .
Submit the job after specifying partition and wall clock criteria. Typically ~2 minutes per system per core is sufficient.
sbatch 001.submit_dock.sh
When docking is completed you will have a separate directory for each system. In each directory will be the input file, output file, and mol2 of docked results. If condition was "Default" and seed was "0", the file will be named:
${pdb_id}/Default_0_scored.mol2
IV.Pose Reproduction Analysis
Next run a script which calculates outcomes. This script is compatible with python 2.
module load py/2.7.15
python calculate_dock6.results.py ${condition}_${seed} ${system_file}
e.g.: python calculate_dock6.results.py Default_0 clean.systems.all
In the below image, the systems "1KIJ","1QCA","2AA2" did not successfully dock. There were 1,279 systems in the list provided. The raw number of systems and percentage for "Success", "Score Fail" and "Sample Fail" are given. Incomplete docked systems are counted as Sample Fails.
-SEE README FILE IN GIT REPO FOR ADDTIONAL DETAILS THAT MAY NOT BE COVERED HERE
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (This tutorial was last updated 02/22/2024)
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>