Difference between revisions of "2025 DOCK tutorial 2 with PDBID 1XMU"
Stonybrook (talk | contribs) (→Grid Generation) |
Stonybrook (talk | contribs) (→Surface Sphere Generation) |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Introduction == | == Introduction == | ||
| − | + | ||
| − | + | DOCK is a molecular docking program, initially based on geometric shape-matching algorithm. It includes key features such as Parameterization of ligands and receptors, Physics-based scoring functions, Ligand orientation and flexibility. These make it valuable for virtual screening. Current DOCK version extends beyond virtual screening to support: Genetic algorithms, Molecular evolution, and Molecular dynamics simulations. DOCK has contributed to major drug discovery breakthroughs, including the identification of fatty acid-binding proteins. Dock online users' manual [[https://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm]] | |
| + | |||
| + | |||
| + | '''Tools''' | ||
| + | |||
| + | i. The virtual screening will be performed using the latest version, DOCK 6.12. | ||
| + | |||
| + | ii. Other essential tools include Chimera, which can be downloaded [[https://www.cgl.ucsf.edu/chimera/]] | ||
| + | |||
| + | iii. A High-Performance Computing (HPC) cluster is required for these calculations. This tutorial will use the Seawulf cluster at Stony Brook University. | ||
| + | |||
| + | iv. If unfamiliar with text editors, use '''vimtutor ''' to learn the basics. | ||
| + | |||
| + | |||
| + | '''System of Interest''' | ||
| + | |||
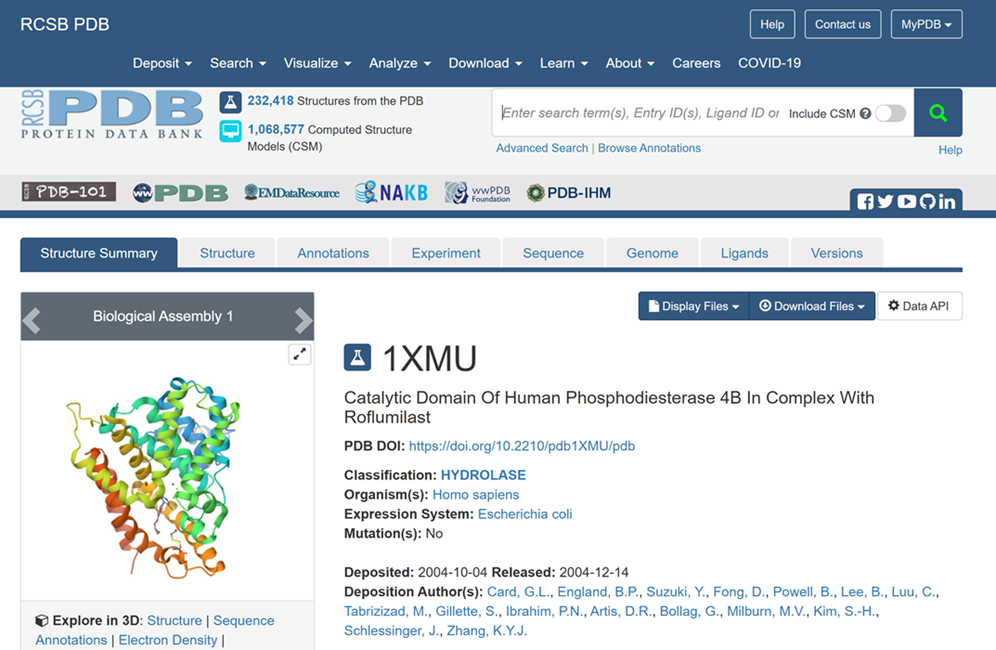
| + | The focus of this tutorial is the Catalytic Domain of Human Phosphodiesterase 4B in Complex with Roflumilast (PDB Code: 1XMU) | ||
| + | |||
| + | i. Visit the RCSB PDB Database [[https://www.rcsb.org/]] and search for 1XMU | ||
| + | |||
| + | ii. Result will be displayed as shown below | ||
| + | |||
| + | [[File:PDB_search_result.png]] | ||
| + | |||
| + | |||
| + | |||
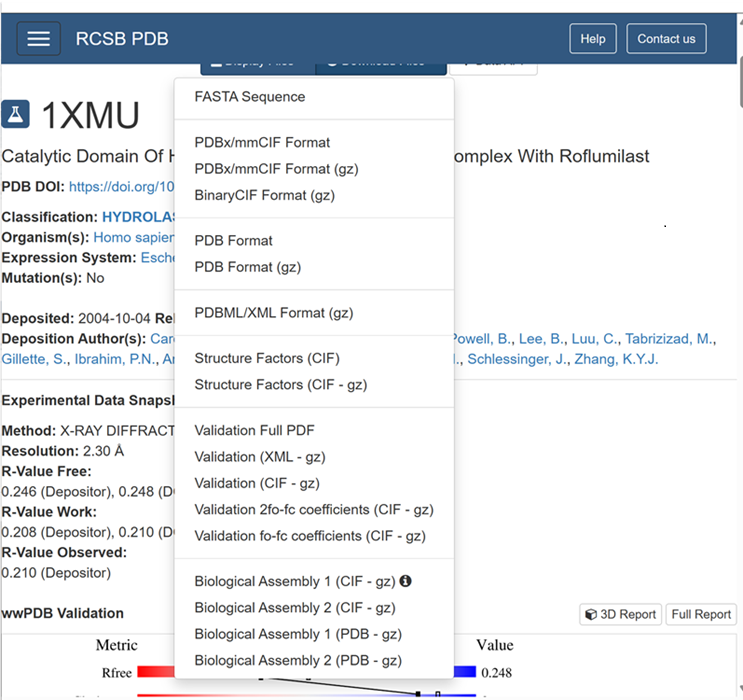
| + | iii. Download the PDB file as (PDB-gz) | ||
| + | |||
| + | |||
| + | [[File:downloadaspdb-gz.png]] | ||
| + | |||
| + | |||
| + | '''Directory Setup''' | ||
| + | |||
| + | On your Seawulf account, set up your working directory for 1XMU by following these steps: | ||
| + | |||
| + | 1. Create the main directory for virtual screening using the following command | ||
| + | |||
| + | mkdir 1XMU_DOCK_VS | ||
| + | |||
| + | 2. Set up the subdirectories inside the 1XMU_DOCK_VS directory by using the following command | ||
| + | |||
| + | mkdir 001.structure 002.surface_spheres 003.gridbox 004.energy_minimization 005.footprint 006.rigid_docking 007.fixed_anchor_docking 008.flex_docking 009.virtual_screening 010.cartesian_minimization 011.rescore | ||
| + | |||
| + | 001.structure | ||
| + | 002.surface_spheres | ||
| + | 003.gridbox | ||
| + | 004.energy_minimization | ||
| + | 005.footprint | ||
| + | 006.rigid_docking | ||
| + | 007.fixed_anchor_docking | ||
| + | 008.flex_docking | ||
| + | 009.virtual_screening | ||
| + | 010.cartesian_minimization | ||
| + | 011.rescore | ||
== Structure Preparation == | == Structure Preparation == | ||
| Line 186: | Line 240: | ||
viii. If successful, the DMS file appears as small dots and align perfectly with the receptor structure. | viii. If successful, the DMS file appears as small dots and align perfectly with the receptor structure. | ||
| − | [[File: | + | [[File:01dms.png]] |
d. Upload dms files to your Seawulf Account | d. Upload dms files to your Seawulf Account | ||
| Line 258: | Line 312: | ||
a. Generate the grid | a. Generate the grid | ||
b. Verify the Process | b. Verify the Process | ||
| + | |||
'''1. Showbox''' | '''1. Showbox''' | ||
| + | |||
'''a. Generate the Binding Site Box''' | '''a. Generate the Binding Site Box''' | ||
| Line 280: | Line 336: | ||
showbox < showbox.in | showbox < showbox.in | ||
| + | |||
'''b. Verify the Process''' | '''b. Verify the Process''' | ||
| Line 326: | Line 383: | ||
i. If successful, three new files named grid.nrg, grid.bmp, and 1XMU_grid.out will appear in your directory | i. If successful, three new files named grid.nrg, grid.bmp, and 1XMU_grid.out will appear in your directory | ||
| + | |||
| + | ii. If these files are missing or the process fails, check the grid output file (grid.out) for error messages | ||
| + | |||
| + | iii. Identify any issues and troubleshoot accordingly | ||
== Energy Minimization == | == Energy Minimization == | ||
Latest revision as of 15:35, 10 March 2025
Contents
Introduction
DOCK is a molecular docking program, initially based on geometric shape-matching algorithm. It includes key features such as Parameterization of ligands and receptors, Physics-based scoring functions, Ligand orientation and flexibility. These make it valuable for virtual screening. Current DOCK version extends beyond virtual screening to support: Genetic algorithms, Molecular evolution, and Molecular dynamics simulations. DOCK has contributed to major drug discovery breakthroughs, including the identification of fatty acid-binding proteins. Dock online users' manual [[1]]
Tools
i. The virtual screening will be performed using the latest version, DOCK 6.12.
ii. Other essential tools include Chimera, which can be downloaded [[2]]
iii. A High-Performance Computing (HPC) cluster is required for these calculations. This tutorial will use the Seawulf cluster at Stony Brook University.
iv. If unfamiliar with text editors, use vimtutor to learn the basics.
System of Interest
The focus of this tutorial is the Catalytic Domain of Human Phosphodiesterase 4B in Complex with Roflumilast (PDB Code: 1XMU)
i. Visit the RCSB PDB Database [[3]] and search for 1XMU
ii. Result will be displayed as shown below
iii. Download the PDB file as (PDB-gz)
Directory Setup
On your Seawulf account, set up your working directory for 1XMU by following these steps:
1. Create the main directory for virtual screening using the following command
mkdir 1XMU_DOCK_VS
2. Set up the subdirectories inside the 1XMU_DOCK_VS directory by using the following command
mkdir 001.structure 002.surface_spheres 003.gridbox 004.energy_minimization 005.footprint 006.rigid_docking 007.fixed_anchor_docking 008.flex_docking 009.virtual_screening 010.cartesian_minimization 011.rescore
001.structure 002.surface_spheres 003.gridbox 004.energy_minimization 005.footprint 006.rigid_docking 007.fixed_anchor_docking 008.flex_docking 009.virtual_screening 010.cartesian_minimization 011.rescore
Structure Preparation
The objectives of this section are:
1. Structural Evaluation
a. Identify any missing loops in the protein structure b. Assess metal coordination atoms for key interactions with the protein
2. Protein Receptor Preparation
a. Generate protein structure in pdb and mol2 format b. Add hydrogen atoms c. Assign appropriate charges d. Save the refined protein structure in mol2 format
3. Ligand Preparation
a. Generate ligand structure in mol2 format b. Add hydrogen atoms c. Assign appropriate charges d. Save the refined ligand structure in mol2 format
4. Alternative method of protein and ligand preparation
a. Using Dock Prep
5. Uploading mol2 Files to your Seawulf Account
a. Transfer via computer terminal using SCP, or b. Transfer via MobaXterm (SFTP)
1. Structural Evaluation
a. Open the downloaded PDB structure in Chimera (In this tutorial, 1XMU). Check for missing loops—these appear as dashed lines in the structure. There are no missing loops in the structure 1XMU, so no further action is needed.
If you find missing loops in your structure, you will need to fix them before proceeding. Refer to (Tutorial 2025 [[4]]) for instructions on fixing missing loops.
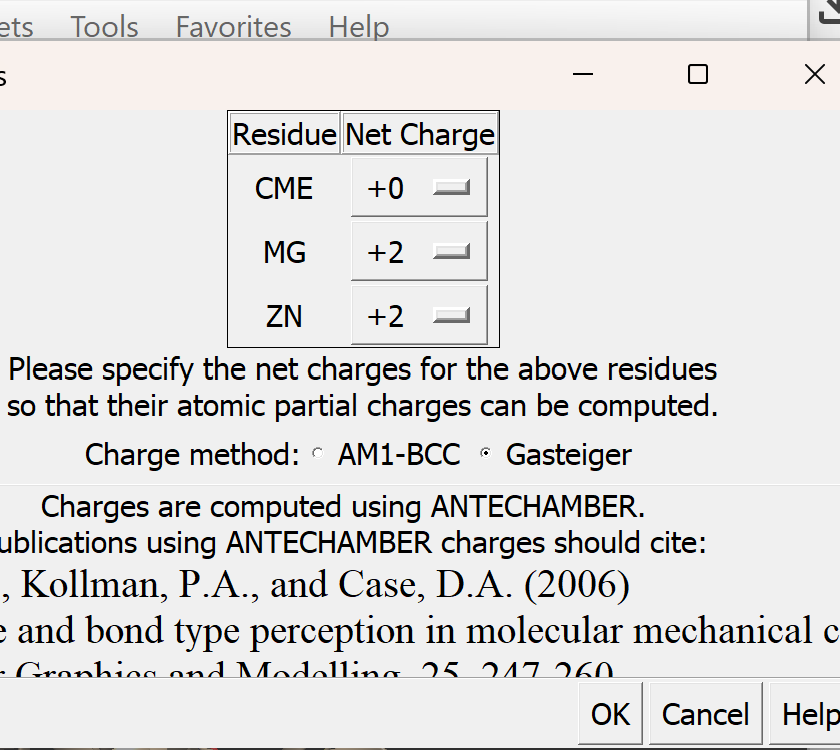
b. In 1XMU, there is metal coordination atoms (Zinc and Magnesium) interacting with the protein. It is important to keep them intact and not delete them during protein preparation
2. Protein Receptor Preparation
a. Generate protein structure in pdb and mol2 format
i. Open the downloaded pdb file (1XMU) in Chimera,
ii. Remove ligand: Go to Select → residue → ligand to highlight the ligand
iii. Then, go to Actions → Atoms/Bonds → Delete
iv. Repeat the process to delete water molecules by selecting HOH instead of ligand
v. Save the protein, Go to File → Save Mol2 and save as 1XMU_Rec_nCH.mol2 ('no' Charge and Hydrogen). Also, save the structure in PDB format as 1XMU_Rec_nCH.pdb.
b. Add Hydrogen atoms
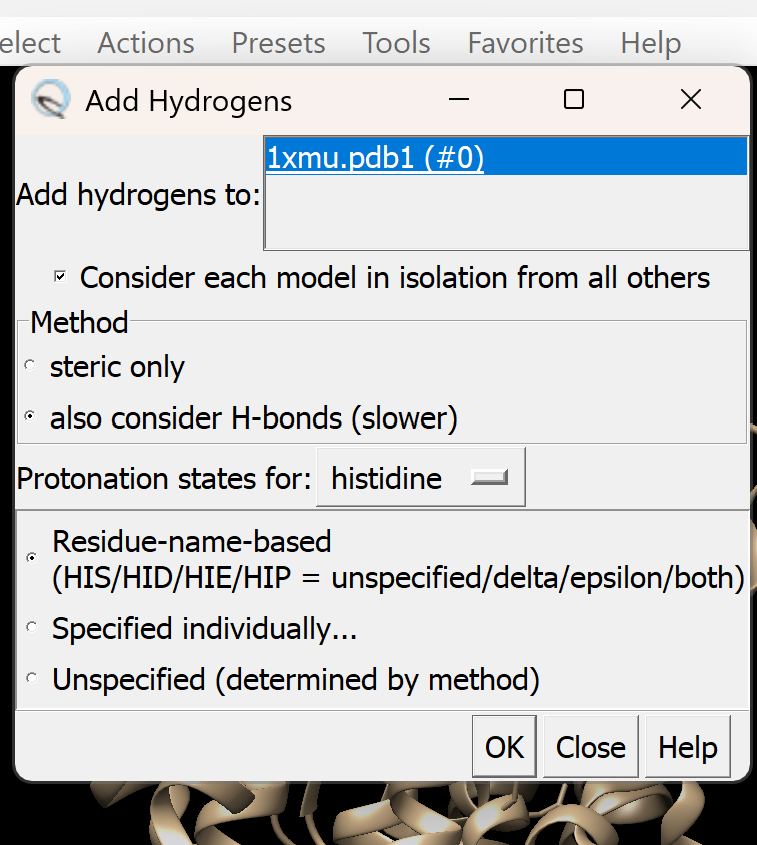
i. Go to Tools → Structure Editing → AddH. A dialogue box like this will appear, adjust as needed
ii. Select OK to proceed. If successful, You will get a confirmation message 'Hydrogens Added'
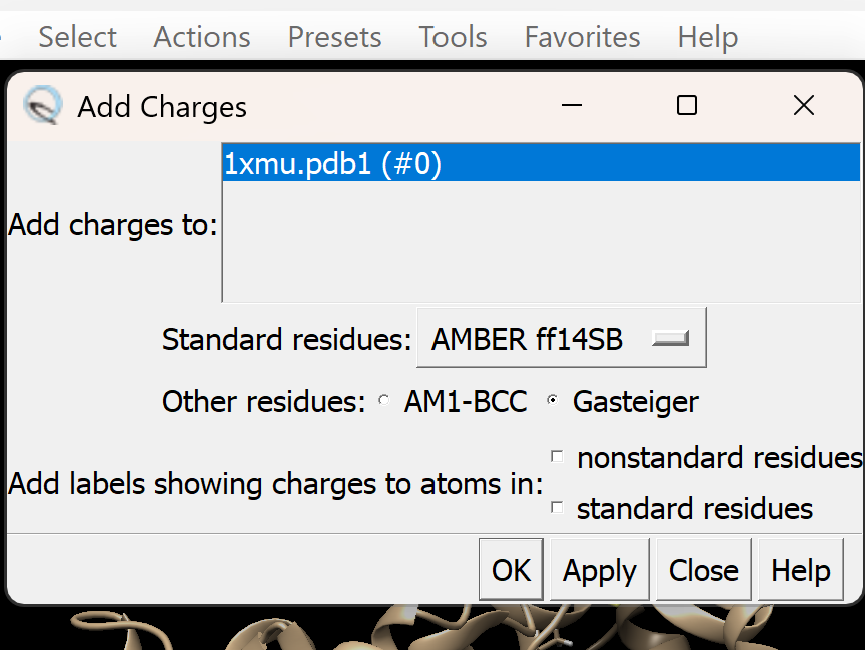
c. Assign appropriate charges
i. Go to Tools → Structure Editing → Add Charge
A dialogue box like this will appear, adjust as needed. For this tutorial, we will use Gasteiger charges. Leave all other settings as they are, then click OK.
ii. A new pop-up menu will appear
iii. Just as before, make sure that Gasteiger is selected. All other defaults are okay.
iv. Select OK. If successful, You will get a confirmation message 'standard charges added'
d. Go to File → Save Mol2. 1XMU_Rec_wCH.mol2 ('with' Charge and Hydrogen)
3. Ligand Preparation
a. Generate ligand structure in mol2 format
i. Open the downloaded pdb file (1XMU) in Chimera,
ii. Remove protein: Go to Select → residue → ligand to highlight the ligand
iii. Then, go to Select → Invert (selected models)to select everything except the ligand
iv. Go to Actions → Atoms/Bonds → Delete
iv. Save the ligand, Go to File → Save Mol2. 1XMU_lig_nCH.mol2 ('no' Charge and Hydrogen)
b. Add Hydrogen atoms
i. Go to Tools → Structure Editing → AddH
ii. The next menu will be the same as when adding hydrogens during receptor preparation. Use the same settings as before
c. Assign appropriate charges
i. Go to Tools → Structure Editing → Add Charge
ii. The next menu will be the same as when adding hydrogens during receptor preparation. Use the same settings as before
Emphasis: For this tutorial, we will use Gasteiger charge
d. Go to File → Save Mol2. 1XMU_lig_wCH.mol2 ('with' Charge and Hydrogen)
4. Alternative Method for Protein and Ligand Preparation
a. Using Dock Prep
This method simplifies preparation by adding hydrogens and assigning charges in a single step.
i. Open the PDB file (1XMU) in Chimera
ii. Isolate the protein and ligand following the steps in Sections 2a and 3a
iii. Go to Tools → Structure Editing → Dock Prep
iv. A pop-up menu will appear – leave all default parameters (as seen below) and click OK
v. The next set of drop-down menus will follow the same process as adding hydrogens and charges manually
vi. The final pop-up menu will be the Save mol2 window
5. Uploading mol2 Files to Your Seawulf Account
a. Transfer via computer terminal using SCP
Use the scp command to transfer files from your local computer to Seawulf: for example,
scp \Users\Documents\1XMU_DOCK_VS\001.structure\1XMU_lig_nCH.mol2 oogunfolakan@milan.seawulf.stonybrook.edu:/gpfs/home/oogunfolakan/1XMU_DOCK_VS/001.structure
NOTE: When using scp via computer terminal, Seawulf automatically pushes DUO authentication to the first registered device i.e you cannot choose which device receives the DUO prompt
b. Transfer via MobaXterm (SFTP)
If you're using MobaXterm, you can upload files directly using the SFTP panel in your SSH browser.
NOTE: Navigate to your target directory (001.structure in this case) on seawulf and drag and drop the files for upload
Surface Sphere Generation
Sphere generation is essential for identifying receptor active sites in DOCK calculations. The objectives of this section are:
1. Write DMS file
a. Generate Receptor Surface b. Generate the DMS file c. Verify the Process d. Upload dms files to your Seawulf Account
2. Run Sphere Generation on Seawulf
a. Generate sphere b. Verify the Process
3. Selecting Active/Binding Site Spheres
a. Run Sphere Selection b. Verify the Process
1. Write DMS file
a. Generate Receptor Surface
i. Open the Receptor File.pdb (without charges and hydrogens) in Chimera
ii. Go to Actions → Surface → Show
iii. The receptor's surface should now be displayed
b. Generate the DMS file
iv. Go to Tools → Structure Editing → Write DMS
v. A menu to save the file as dms will appear (1XMU_surface.dms)
c. Verify the Process
vi. First of all, Open the 1XMU receptor file.pdb in Chimera
vii. Then, open the 1XMU_spheres.dms file.
viii. If successful, the DMS file appears as small dots and align perfectly with the receptor structure.
d. Upload dms files to your Seawulf Account
ix. On your Seawulf account, move to the 002.surface_spheres directory
x. Use either SCP or MobaXterm (SFTP) to upload 1XMU_spheres.dms as discussed earlier.
2. Run Sphere Generation on Seawulf
a. Generate sphere
i. Create an input file using the command vi INSPH
ii. Type in the following code (adjust your file names accordingly)
1XMU_spheres.dms R X 0.0 4.0 1.4 1XMU.sph
iii. Save the file and exit
iv. Run sphgen to generate spheres using the command
sphgen -i INSPH -o OUTSPH
b. Verify the Process
v. If sphgen runs successfully, the output file 1XMU.sph should appear in your directory
vi. Launch Chimera and open the generated sphere file (1XMU.sph). Then, open the receptor PDB file (1XMU_Rec_nCH.pdb)
vii. The spheres should perfectly overlay with the receptor structure
3. Selecting Active/Binding Site Spheres
DOCK6 provides a program called sphere_selector, which isolates spheres specifically at the receptor's active site. There is need to specify the distance (in this tutorial, 10.0 Å) in the command to ensure that sphere_selector selects spheres within distance of the ligand, accurately defining the binding pocket.
a. Run Sphere Selection
i. Move to the 002.surface_spheres directory on your Seawulf account
ii. Run Sphere Selection using the command
sphere_selector 1XMU.sph ../001.structure/1XMU_lig_wCH.mol2 10.0
b. Verify the Process
iii. If successful, a new file named selected_spheres.sph will appear in your directory
iv. Open Chimera and load the selected_spheres. Then, open the original 1XMU.pdb in the same session
v. If successful, the spheres should be positioned where the ligand is, confirming that the binding site has been correctly identified
Grid Generation
Calculating interactions between the ligand and the entire protein surface is computationally expensive. To focus calculations on the active site, we use DOCK6’s showbox to create a box enclosing the binding spheres. This helps DOCK generate a grid for interaction energy calculations. In this tutorial, this box extends 8.0 Å from the binding spheres. The objectives of this section are:
1. Showbox
a. Generate the Binding Site Box b. Verify the Process
2. Generating the Grid
a. Generate the grid b. Verify the Process
1. Showbox
a. Generate the Binding Site Box
i. Create an input file using the command
vi showbox.in
ii. Type in the following code (adjust your file names accordingly)
Y 8.0 ../002.surface_spheres/selected_spheres.sph 1 1XMU.box.pdb
iii. Save the file and exit
iv. Generate the box using the command
showbox < showbox.in
b. Verify the Process
i. If successful, a new file named 1XMU.box.pdb will appear in your directory
2. Generating the Grid
a. Generate the grid
i. Create an input file using the command
vi grid.in
ii. Type in the following code (adjust your file names accordingly)
allow_non_integral_charges no compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../001.structure/1XMU_Rec_wCH.mol2 box_file 1XMU.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.12/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
iii. Save the file and exit
iv. Generate the grid using the command
grid -i grid.in -o 1XMU_grid.out
b. Verify the Process
i. If successful, three new files named grid.nrg, grid.bmp, and 1XMU_grid.out will appear in your directory
ii. If these files are missing or the process fails, check the grid output file (grid.out) for error messages
iii. Identify any issues and troubleshoot accordingly