Difference between revisions of "2008 DOCK tutorial with Gleevec"
| (3 intermediate revisions by the same user not shown) | |||
| Line 114: | Line 114: | ||
Log_P: 3.08 | Log_P: 3.08 | ||
| − | |||
| + | |||
| + | [[Image:Gleevec.correct.png|thumb|400px|right|Gleevec should most likely be neutral for our purposes . See http://en.wikipedia.org/wiki/Imatinib for more details]] | ||
| + | |||
| + | '''CORRECTION''': Gleevec should not have a +2 net charge. | ||
| + | |||
| + | In chimera, add hydrogens. Then, if present, remove the hydrogens from the two nitrogens in Piperazine ring. Next add charges to the molecule and the net charge should be zero. | ||
| + | |||
| + | Note the protination states of small molecules are often difficult to determine and are extremely important to binding. Some time programs do not predict the correct protonation state like above and the user should varify or make an educated guess at the protonation state do not asume that the protonation and hybridization are correct. | ||
| + | |||
| + | |||
| + | |||
| + | |||
== Generating the Receptor Molecular Surface == | == Generating the Receptor Molecular Surface == | ||
Latest revision as of 09:30, 19 May 2011
For additional Rizzo Lab tutorials see DOCK Tutorials.
Contents
About DOCK
DOCK was developed by Irwin D. "Tack" Kuntz, Jr., PhD and colleagues at UCSF. Please see the webpage at UCSF DOCK.
DOCK is a molecular docking program used in drug discovery. This program, given a protein active site and a small molecule, tries to predict the correct binding mode of the small molecule in the active site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug.
Installing DOCK
- Unzip or copy the DOCK distribution to a convenient folder. e.g. /home/sudipto/dock6
- Go to dock6/install, and run ./configure gnu
- Type 'make'. A lot of compilation messages should scroll by the screen, with occasional warnings.
- If everything went well, you should get a message which looks like
Installation of DOCK v6.2dev is complete at Mon Feb 4 14:47:28 EST 2008.
- The DOCK executable should be dock6/bin/dock6
- Update your .cshrc (or equivalent) file to contain path to DOCK bin directory. This can be done where other program variables have been set.
### DOCK ### set path = ( $path $AMS536/dock6/bin)
You need to resource your .cshrc file
source .cshrc
Docking in Gleevec
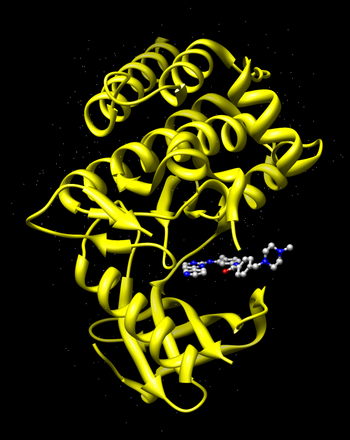
Go to the RCSB Protein Databank and search for Gleevec.You should find the PDB code 1XBB named "Crystal structure of the syk tyrosine kinase domain with Gleevec".
If not, use the link http://www.rcsb.org/pdb/explore.do?structureId=1XBB
Select Download Files in the left window pane. Select "Biological Unit gz". This will give you the biological unit file compressed with gzip. If you can not find it, use the link http://www.rcsb.org/pdb/files/1xbb.pdb1.gz
Download the gzip file and locate it on your desktop. Uncompress the file with the command
gzip -d 1xbb.pdb1.gz
You should now have the file 1xbb.pdb1 Open the file in vi and look at the contents.
It will have a header that looks like this:
HEADER TRANSFERASE 30-AUG-04 XXXX TITLE CRYSTAL STRUCTURE OF THE SYK TYROSINE KINASE DOMAIN WITH TITLE 2 GLEEVEC COMPND Tyrosine-protein kinase SYK (E.C.2.7.1.112) KEYWDS GLEEVEC, STI-571, IMATINIB, SYK, SPLEEN TYPROSINE KINASE, KEYWDS 2 ACTIVE CONFORMATION, STRUCTURAL GENOMICS, STRUCTURAL GENOMIX EXPDTA X-RAY DIFFRACTION AUTHOR V.L.NIENABER, S.ATWELL, J.M.ADAMS, J.BADGER, M.D.BUCHANAN, AUTHOR 2 I.K.FEIL, K.J.FRONING, X.GAO, J.HENDLE, K.KEEGAN, B.C.LEON, AUTHOR 3 H.J.MULLER-DEICKMANN, B.W.NOLAND, K.POST, K.R.RAJASHANKAR, AUTHOR 4 A.RAMOS, M.RUSSELL, S.K.BURLEY, S.G.BUCHANAN JRNL AUTH S.ATWELL, J.M.ADAMS, J.BADGER, M.D.BUCHANAN, JRNL AUTH 2 I.K.FEIL, K.J.FRONING, X.GAO, J.HENDLE, K.KEEGAN, JRNL AUTH 3 B.C.LEON, H.J.MULLER-DEICKMANN, V.L.NIENABER, JRNL AUTH 4 B.W.NOLAND, K.POST, K.R.RAJASHANKAR, A.RAMOS, JRNL AUTH 5 M.RUSSELL, S.K.BURLEY, S.G.BUCHANAN JRNL TITL A NOVEL MODE OF GLEEVEC BINDING IS REVEALED BY THE JRNL TITL 2 STRUCTURE OF SPLEEN TYROSINE KINASE JRNL REF J.BIOL.CHEM. V. 279 55827 2004 JRNL REFN ASTM JBCHA3 US ISSN 1083-351X
Scroll past the SEQRES entries to see the section describing the ligand
HETNAM 2 STI PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE HETSYN STI STI-571 FORMUL 2 STI C29 H31 N7 O
The first line contains the IUPAC name of the ligand, followed by the molecule 'synonym' used in this file for the ligand. The third line describes the molecular formula of the ligand. Note the number of hydrogen atoms here. It is often useful to verify that the correct number of hydrogen atoms have been added during protonation.
Receptor Preparation with Chimera
Open the file 1xbb.pdb1 with Chimera. Examine the PDB file, observe the binding site and the ligand bound to it.
Now select and delete the ligand Gleevec from the complex.
Go to Actions -> Atoms/Bonds -> Delete to delete the ligand molecule.
Now go to Tools->Structure Editing->Dock Prep. This starts the DockPrep module of Chimera. During DockPrep, it will ask for the histidine protonation. In this select 'Unspecified determined by method'.
After DockPrep has run, save the charged receptor file in the .mol2 format (say 'receptor_charged.mol2). (Go to -> File|Save Mol2)
In order to generate the molecular surface of the receptor, one has to use the 'dms' program. For this the Hydrogens of the receptor needs to be removed.
(Select| Chemistry |Element | H and then Actions| Atoms/Bonds |delete
Now save the mutated receptor in .pdb format (say receptor_noH.pdb). (Go to -> File|Save PDB)
For more details about DockPrep, use http://www.cgl.ucsf.edu/chimera/1.2199/docs/ContributedSoftware/dockprep/dockprep.html
Ligand Preparation in Chimera

Close the previous session in Chimera and reopen the original pdb file '1XBB.pdb1' in Chimera.
Now delete the receptor to get only the ligand. (Select| Structure | Ligand. --> Select | Invert all models --> Actions |Atoms/Bonds |Delete)
To prepare the ligand, select the ligand and add hydrogen and charge using Tools | Structure Editing |Add H and similarly Add Charges. The formal charge of the ligand here is +2. Verify that the hydrogens have been added correctly as in the diagram.
Save the molecule in .mol2 format (say 'ligand_charged.mol2).
Molecular Descriptors for Gleevec: Rotatable_Bonds: 7 Heavy_Atoms: 37 HBond_donors: 2 HBond_acceptors: 5 Molecular_Weight: 493.61 Log_P: 3.08
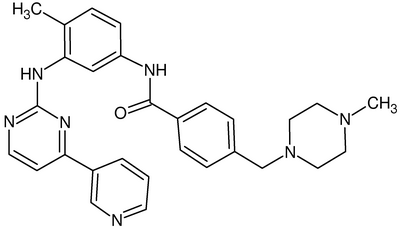
CORRECTION: Gleevec should not have a +2 net charge.
In chimera, add hydrogens. Then, if present, remove the hydrogens from the two nitrogens in Piperazine ring. Next add charges to the molecule and the net charge should be zero.
Note the protination states of small molecules are often difficult to determine and are extremely important to binding. Some time programs do not predict the correct protonation state like above and the user should varify or make an educated guess at the protonation state do not asume that the protonation and hybridization are correct.
Generating the Receptor Molecular Surface
This is done by using the 'dms' program and the mutated receptor file with hydrogens stripped off.
/home/sudipto/dms_program receptor_noH.pdb -n -w 1.4 -v -o receptor.ms
The options associated with the dms program are
-a use all atoms, not just amino acids -d change density of points -g send messages to file -i calculate only surface for specified atoms -n calculate normals for surface points -w change probe radius -v verbose -o specify output file name (required)
Generate Spheres
1. Run sphgen on the command line using the sample input file called INSPH. Note, sphgen requires that the input file be calaled INSPH. The output file is always called OUTSPH. Note, if any of the output files (OUTSPH) or any of the temp files (temp1.ms, temp2.sph or temp3.atc,) exist, a core dump will occur. Just delete all these files and start again). SPHGEN takes a few minutes to run.
sphgen -i INSPH -o OUTSPH
Here is a sample output OUTSPH file.
density type = X reading rec.ms type R # of atoms = 2317 # of surf pts = 64657 finding spheres for rec.ms dotlim = 0.000 radmax = 4.000 Minimum radius of acceptable spheres? 1.39999998 output to rec.sph clustering is complete 32 clusters
2. Select spheres in the proximity of the binding site:
sphere_selector takes as input receptor.sph which that was generated by sphgen, a .mol2 file specifying the ligand binding site and the distance (in angstroms) from the ligand.
sphere_selector receptor.sph ligand_charged.mol2 10.0
3. Convert spheres to PDB format using showsphere program so we can visualize the spheres in the context of the receptor and the original ligand
showsphere <showsph.in
The file selected_spheres.pdb (as defined in showsp.in) is generated after running showsphere.
4. View results
Based on the showsph.in file, this will generate a pdb file selected_spheres.pdb.
This section will use command line mode of Chimera. After starting Chimera, use Tools->General Controls->Command Line Adjust the side view to get entire protein in view (Tools->Viewing Controls->Side View
open selected_spheres.pdb open receptor_charged.mol2 open ligand_charged.mol2 surface #0 ~disp #0 surface #1 ~disp #1 surface #2 ~disp #2 surftrans 50 #0
This will open the spheres, receptor and ligand and turns on the surface display and turns off the atom/bond display. The transparency of the spheres is set to 50%
Generate Grid
Generate grid box.
showbox < showbox.in
After running showbox we can view the box that the grid will be placed in.
Start chimera
Open the receptor, spheres from sphgen and the box from showbox.
In Chimera, select
File->Open-> and select your receptor molecule (ie receptor_charged.mol2) File->Open-> and select your box (ie rec_box.pdb) File->Open-> and select your spheres (ie selected_spheres.pdb)
Make the receptor a surface representation. Select->Chain->A Action->Surface->Show Action->Atoms/Bonds->Hide
Select your grid box and enlarge the lines.
Select->Residue->Box Actions->Atoms/Bonds->Wire Width->4
Increase size on the selected spheres. Select->Chain->(No id)->Selected Spheres Action->Atoms/Bonds->Ball+Stick Tools->General Controls->Model Panel
Select the selected_spheres model by clicking once on the name.
Select the 'Attributes' button on the right. Set the ball scale to .5.
The following figure shows the surface representation of the receptor molecule in red. The spheres that occupy the nooks and crannies represent potential sites that heavy atom molecules can potentially occupy.
One can also run the following commands in command line mode.
open receptor_charged.mol2 open rec_box.pdb open selected_spheres.pdb surface #0 ~disp #0 linewidth 4 #1 repr bs #2 setattr ballScale .5 #2
Generate grid
grid -i grid.in
This generates grid.nrg (energy scoring file) and grid.bmp (bump grid) files. The grid computation creates an energy grid grid.nrg and a bump grid grid.bmp. This computation should take about 5 mins to finish.
If you need to run grid again, please ensure that grid.bmp and grid.nrg files are removed.
Perform Docking
There are two kinds of docking options : Rigid Ligand Docking and Flexible Ligand Docking.
Rigid Ligand Docking In this the ligand is held rigid during orientation.
dock6 -i rigid.in -o rigid.out
This generates the following files:
rigid.out which gives a summary of the run,
rigid_scored.mol2 which gives the MOL2 file containing the geometric coordinates as well as the summary of interaction energies of binding for the docked ligand poses in the order of their grid score. The best pose would have the most favourable grid score. The following shows a sample view of the details of interaction energies generated in the mol2 file.
########## Name: receptor.pdb ########## RMSD: 1.01108 ########## Grid Score: -52.555294 ########## vdw: -37.118080 ########## es: -15.437213
To visualize the docking result, open rigid_scored.mol2 in Chimera and simultaneously load the receptor.pdb and ligand_charged.mol2 files.
Flexible Ligand Docking
In this type of docking, the ligand is allowed to be flexible by applying an 'anchor and grow' algorithm.
dock6 -i flex.in -o flex.out
This generates the following files:
flex.out giving the summary of the run.
flex_scored.mol2 which gives the geometric coordinates and the summary of the interaction energies of binding for the docked ligand poses in order of their grid score.