Difference between revisions of "2012 DOCK tutorial with Streptavidin"
Stonybrook (talk | contribs) (→Virtual Screening Protocol) |
(→Barbara) |
||
| (62 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | For additional Rizzo Lab tutorials see [[DOCK Tutorials]]. | + | For additional Rizzo Lab tutorials see [[DOCK Tutorials]]. Use this link [http://www.unilang.org/wiki/index.php/Wiki_markup Wiki Markup] as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2012. |
| − | |||
| − | Use this link [http://www.unilang.org/wiki/index.php/Wiki_markup Wiki Markup] as a reference for editing the wiki. | ||
==I. Introduction== | ==I. Introduction== | ||
| Line 45: | Line 43: | ||
===Preparing the Ligand and Enzyme in Chimera=== | ===Preparing the Ligand and Enzyme in Chimera=== | ||
| − | If you are on "herbie" you can access Chimera directly by typing in '''Chimera''' at the prompt. Otherwise, download Chimera to your desktop and obtain your protein/nucleic acid complex of interest '''Fetch''' (by ID). | + | If you are on "herbie" you can access Chimera directly by typing in '''Chimera''' at the prompt. Otherwise, download Chimera to your desktop and obtain your protein/nucleic acid complex of interest using '''Fetch''' (by ID). |
<u> Open Your Protein of Interest and Delete Unwanted Molecules: </u> | <u> Open Your Protein of Interest and Delete Unwanted Molecules: </u> | ||
| Line 115: | Line 113: | ||
1DF8.receptor.sph #clustered spheres file that we want to generate | 1DF8.receptor.sph #clustered spheres file that we want to generate | ||
| − | Save the INSPH file. Then use this command to generate spheres file: | + | Save the INSPH file. Then use this command to generate the spheres file: |
sphgen -i INSPH -o OUTSPH | sphgen -i INSPH -o OUTSPH | ||
| Line 135: | Line 133: | ||
clustering is complete 28 clusters | clustering is complete 28 clusters | ||
| − | You can also open the spheres file that we generated in this step <b>(1DF8_receptor.sph)</b>. This file contains detailed information | + | You can also open the spheres file that we generated in this step <b>(1DF8_receptor.sph)</b>. This file contains detailed information about the spheres, which are divided into 28 clusters. Cluster 0 in the end of the spheres file is a combination of all the clusters. |
In order to visualize the generated spheres, you can use a program called <b>showsphere</b>. Showsphere is an interactive program. In the command line, simply type | In order to visualize the generated spheres, you can use a program called <b>showsphere</b>. Showsphere is an interactive program. In the command line, simply type | ||
| Line 141: | Line 139: | ||
showsphere | showsphere | ||
| − | You will be asked | + | You will be asked the following questions: |
Enter name of sphere cluster file: | Enter name of sphere cluster file: | ||
| Line 165: | Line 163: | ||
showshpere < showsphere.in | showshpere < showsphere.in | ||
| − | After generating the pdb files by showsphere, you can visualize each cluster by UCSF Chimera. For example, open <b>1DF8.output_1.pdb</b> by Chimera, then choose <b>Actions->Atoms/Bonds->sphere</b>, you will be able to see the spheres in cluster 1. You can also open the receptor file <b>(1DF8.receptor.mol2)</b> at the same time, then choose <b>Presets->Interactive 3</b>(hydrophobicity surface), then again choose <b>Actions->Atoms/Bonds->sphere</b>, you will be able to see what | + | After generating the pdb files by showsphere, you can visualize each cluster by UCSF Chimera. For example, open <b>1DF8.output_1.pdb</b> by Chimera, then choose <b>Actions->Atoms/Bonds->sphere</b>, you will be able to see the spheres in cluster 1. You can also open the receptor file <b>(1DF8.receptor.mol2)</b> at the same time, then choose <b>Presets->Interactive 3</b>(hydrophobicity surface), then again choose <b>Actions->Atoms/Bonds->sphere</b>, you will be able to see what the spheres in cluster 1 look like on the enzyme surface. |
[[Image:2012_DOCK_Tutorial_1DF8_spheres.png|thumb|center|375px|2012_DOCK_Tutorial_1DF8_spheres]] | [[Image:2012_DOCK_Tutorial_1DF8_spheres.png|thumb|center|375px|2012_DOCK_Tutorial_1DF8_spheres]] | ||
| Line 173: | Line 171: | ||
sphere_selector 1DF8.receptor.sph ../01-dockprep/1DF8.ligand.mol2 10.0 | sphere_selector 1DF8.receptor.sph ../01-dockprep/1DF8.ligand.mol2 10.0 | ||
| − | Sphere_selector filters the output from <b>sphgen</b>, selecting all spheres within a user-defined radius of a target molecule. In this case, we selected the spheres | + | Sphere_selector filters the output from <b>sphgen</b>, selecting all spheres within a user-defined radius of a target molecule. In this case, we selected the spheres within 10 angstroms of our ligand. A file called <b>selected_spheres.sph</b> should be created, showing the spheres that are selected. You can again visualize it by <b>showsphere</b>. You can also change the radius (to 8 angstroms or 6 angstroms) or manually edit the file selected_spheres.sph so that you can select the spheres you want. In this tutorial, spheres within 6 angstroms of the ligand are used for the next step (with one sphere which does not belong to the active site being manually deleted). |
[[Image:2012_DOCK_Tutorial_1DF8_surface_spheres_10 angstroms.png|thumb|375px|left|2012_DOCK_Tutorial_1DF8_surface_spheres_10 angstroms]] | [[Image:2012_DOCK_Tutorial_1DF8_surface_spheres_10 angstroms.png|thumb|375px|left|2012_DOCK_Tutorial_1DF8_surface_spheres_10 angstroms]] | ||
| + | |||
[[Image:2012_DOCK_Tutorial_1DF8_surface_spheres_6 angstroms.png|thumb|375px|center|2012_DOCK_Tutorial_1DF8_surface_spheres_6 angstroms]]<BR> | [[Image:2012_DOCK_Tutorial_1DF8_surface_spheres_6 angstroms.png|thumb|375px|center|2012_DOCK_Tutorial_1DF8_surface_spheres_6 angstroms]]<BR> | ||
| Line 181: | Line 180: | ||
===Box=== | ===Box=== | ||
| − | In order to speed up docking calculations, DOCK generates a fine grid | + | In order to speed up docking calculations, DOCK generates a fine grid. At each point in the grid, electrostatic and the VDW probes' energies are precomputed. The energies are computed using a molecular force field. To determine the dimensions of the grid, however, we first generate a box that contains the outer boundaries for the grid calculation. The dimensions and location of the box can be determined using a program called '''showbox'''. |
First create a directory where you will place the grid files. | First create a directory where you will place the grid files. | ||
| Line 212: | Line 211: | ||
===Grid=== | ===Grid=== | ||
| − | Now let's generate a grid within our box. We will use the energy scoring method to generate a grid, resulting in three additional files with extensions '''*.nrg''', '''*.bmp''', and '''*.out'''. The '''*.nrg''' file contains the energy scoring, '''*.bmp''' contains the size, position and grid spacing and determines whether there are any | + | Now let's generate a grid within our box. We will use the energy scoring method to generate a grid, resulting in three additional files with extensions '''*.nrg''', '''*.bmp''', and '''*.out'''. The '''*.nrg''' file contains the energy scoring, '''*.bmp''' contains the size, position and grid spacing and determines whether there are any overlaps with receptor atoms. |
To generate the grid we will use the grid program. This program can either be used interactively, or an input file can be fed in, just like the '''showbox''' program. | To generate the grid we will use the grid program. This program can either be used interactively, or an input file can be fed in, just like the '''showbox''' program. | ||
| Line 304: | Line 303: | ||
orient_ligand yes | orient_ligand yes | ||
| − | The "orient_ligand" option tells the program whether to try orientations different from the pose in the original .mol2 file. Note that because of the change, additional questions are asked by the program. For simplicity we keep most of the answers as default. File paths | + | The "orient_ligand" option tells the program whether to try orientations different from the pose in the original .mol2 file. Note that because of the change, additional questions are asked by the program. For simplicity we keep most of the answers as default. File paths that need to be specified are listed below: |
receptor_site_file [receptor.sph] ():../02-surface-spheres/selected_spheres_06A.sph | receptor_site_file [receptor.sph] ():../02-surface-spheres/selected_spheres_06A.sph | ||
| Line 311: | Line 310: | ||
flex_drive_file [flex_drive.tbl] ():../03-grid/flex_drive.tbl | flex_drive_file [flex_drive.tbl] ():../03-grid/flex_drive.tbl | ||
| − | The receptor_site_file should be the selected spheres file (.sph) generated in a previous step (02 surface and spheres), according which the program orients the ligand. | + | The receptor_site_file should be the selected spheres file (.sph) generated in a previous step (02 surface and spheres), according to which the program orients the ligand. |
The vdw_defn_file instructs the dock6 program to use the Van der Waals parameter sets from the AMBER force field. | The vdw_defn_file instructs the dock6 program to use the Van der Waals parameter sets from the AMBER force field. | ||
| Line 328: | Line 327: | ||
num_scored_conformers 10 | num_scored_conformers 10 | ||
| − | Again, further questions will be asked when you run the program. Keep most answers as default. Paths requiring specification are listed below: | + | Again, further questions will be asked when you run the program. Keep most answers as the default values. Paths requiring specification are listed below: |
grid_score_grid_prefix [grid] ():../03-grid/1DF8.grid | grid_score_grid_prefix [grid] ():../03-grid/1DF8.grid | ||
| Line 352: | Line 351: | ||
flexible_ligand yes | flexible_ligand yes | ||
| − | Further questions will be asked during the run. Keep most of the answers as default except for: | + | Further questions will be asked during the run. Keep most of the answers as the default values except for: |
simplex_grow_max_iterations [20] ():500 | simplex_grow_max_iterations [20] ():500 | ||
| − | The simplex_grow_max_iteration specifies the maximum | + | The simplex_grow_max_iteration specifies the maximum number of iterations per cycle per growth step. |
Notice that the run is significantly slower this time. Again the best pose generated (-64.8) is shown in blue. The grid score improved significantly but the RMSD did not change much (0.79) | Notice that the run is significantly slower this time. Again the best pose generated (-64.8) is shown in blue. The grid score improved significantly but the RMSD did not change much (0.79) | ||
| Line 366: | Line 365: | ||
bump_filter yes | bump_filter yes | ||
| − | The bump_filter option filters out conformations that cause | + | The bump_filter option filters out conformations that cause clashes between atoms. |
| − | Further questions will be asked during the run. | + | Further questions will be asked during the run. Keep answers as the default values. Specify the following path: |
bump_grid_prefix [grid] ():../03-grid/1DF8.grid | bump_grid_prefix [grid] ():../03-grid/1DF8.grid | ||
| Line 480: | Line 479: | ||
<b>Virtual screening</b> is a widely used method in computational drug design. It searches large libraries of chemical compounds to identify favorable structures that bind to a target molecule. To perform virtual screening, we use <b>ligands.mol2</b>, a mol2 file which contains 101 small molecules to be the virtual library. The computational cost is reasonable for a quick search. Usually we use larger molecule set from chemical database, such as <b>ZINC</b>(http://zinc.docking.org/). | <b>Virtual screening</b> is a widely used method in computational drug design. It searches large libraries of chemical compounds to identify favorable structures that bind to a target molecule. To perform virtual screening, we use <b>ligands.mol2</b>, a mol2 file which contains 101 small molecules to be the virtual library. The computational cost is reasonable for a quick search. Usually we use larger molecule set from chemical database, such as <b>ZINC</b>(http://zinc.docking.org/). | ||
| − | Since the computational cost of virtual screening is much higher than | + | Since the computational cost of virtual screening is much higher than single-ligand docking, it is better for us to run it on Seawulf. We need to compress the whole <b>DOCK-Tutorial</b> directory, copy it to Seawulf and compress it. |
tar -cvf DOCK-Tutorial.tar DOCK-Tutorial/ | tar -cvf DOCK-Tutorial.tar DOCK-Tutorial/ | ||
scp DOCK-Tutorial sw:/nfs/user03/usrname/AMS536 | scp DOCK-Tutorial sw:/nfs/user03/usrname/AMS536 | ||
| Line 511: | Line 510: | ||
> cluster_rmsd_threshold 0.2 | > cluster_rmsd_threshold 0.2 | ||
| − | <b>num_scored_conformers 1 -> 10</b> | + | <b>num_scored_conformers: 1 -> 10</b> In virtual screening, we only need the most favorable pose of each candidate molecule and compare them.<br> |
| − | <b>cluster_conformations yes -> no</b> | + | <b>cluster_conformations: yes -> no</b> Slightly different conformations are not clustered together. They are treated as different conformations in the docking process. |
| + | |||
| + | In order to generate different search spaces, we can modify some other parameters. | ||
| + | |||
| + | <b>max_orientations:</b> The maximal number of anchor orientations that will be generated.<br> | ||
| + | <b>min_anchor_size:</b> The minimum number of atoms for an anchor.<br> | ||
| + | <b>pruning_use_clustering:</b> Pruning conformations during the clustering process.<br> | ||
| + | <b>use_internal_energy:</b> Using repulsive VDM to avoid internal clashes.<br> | ||
| + | |||
| + | Parallel computing can reduce the running time of virtual screening. Here is our job submission script <b>sub.virtual_screen.csh</b>. | ||
| + | #! /bin/tcsh | ||
| + | #PBS -l nodes=4:ppn=2 | ||
| + | #PBS -l walltime=01:00:00 | ||
| + | #PBS -o zzz.qsub.out | ||
| + | #PBS -j oe | ||
| + | #PBS -V | ||
| + | set nprocs = `wc -l $PBS_NODEFILE | awk '{print $1}'` | ||
| + | echo "Running on ${nprocs} processors" | ||
| + | echo "" | ||
| + | echo "processor list are:" | ||
| + | cat $PBS_NODEFILE | ||
| + | cd ~/AMS536/DOCK_Tutorial/06-virtualscreen | ||
| + | mpirun -np $nprocs dock6.mpi -i vs.in -o vs.out | ||
| + | Finally, we can submit the job and perform virtual screening. | ||
| + | qsub sub.virtual_screen.csh | ||
===Virtual Screening Results=== | ===Virtual Screening Results=== | ||
| Line 606: | Line 629: | ||
===Parallel Virtual Screen=== | ===Parallel Virtual Screen=== | ||
| − | ==VIII. Frequently Encountered Problems== | + | ==VIII. Molecular Footprints== |
| + | |||
| + | ===Preparation (minimization) of Reference Molecule=== | ||
| + | The footprint of the reference molecule guides the selection of molecules/poses in the footprint rescoring method, therefore careful preparation of this reference molecule is necessary. Typically the reference molecule will be an endogenous substrate or known inhibitor from an x-ray crystallographic structure. | ||
| + | |||
| + | Preferably, the molecule should be prepared with hydrogens and charges using AMBER and subsequently converted to mol2 format using the 'top2mol2' command. This will provide you with a 'monotonic' mol2 file -- meaning that all the hydrogens will be listed with the corresponding protein residue, rather than separately at the end of the file (like Chimera's 'DockPrep' tool does). | ||
| + | |||
| + | An alternative way to generate the "monotonic" receptor file is to open the receptor.mol2 file with Chimera, save as pdb file, reopen the pdb file and go through DockPrep, and again save as a mol2 file. In this case, the new mol2 file will be monotonic. | ||
| + | |||
| + | Once the molecule is prepared, it should be minimized in the receptor using a tethered minimization. This is especially important if the crystal structure was refined without hydrogens (typically hydrogens are not used in refinement unless the resolution is ~1 Angstrom or better). Minimization is needed to help alleviate any steric clashes involving the newly added hydrogens (which would otherwise disrupt your footprint results). | ||
| + | |||
| + | An example input file for use on seawulf (using dock6.5): | ||
| + | |||
| + | ligand_atom_file ligand.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol no | ||
| + | use_database_filter no | ||
| + | orient_ligand no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | flexible_ligand no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary no | ||
| + | grid_score_secondary no | ||
| + | dock3.5_score_primary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_primary yes | ||
| + | continuous_score_secondary no | ||
| + | cont_score_rec_filename receptor.mol2 | ||
| + | cont_score_att_exp 6 | ||
| + | cont_score_rep_exp 12 | ||
| + | cont_score_rep_rad_scale 1 | ||
| + | cont_score_use_dist_dep_dielectric yes | ||
| + | cont_score_dielectric 4.0 | ||
| + | cont_score_vdw_scale 1 | ||
| + | cont_score_es_scale 1 | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | simplex_max_iterations 1000 | ||
| + | simplex_tors_premin_iterations 0 | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min yes | ||
| + | simplex_coefficient_restraint 10.0 | ||
| + | atom_model all | ||
| + | vdw_defn_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/flex.defn | ||
| + | flex_drive_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix xtalmin | ||
| + | write_orientations no | ||
| + | num_scored_conformers 1 | ||
| + | rank_ligands no | ||
| + | |||
| + | ===Molecular footprints comparisons=== | ||
| + | |||
| + | After rescoring of the molecules by footprint-based score, typically three output files will be generated: a mol2 file showing the ligands information, a footprint.txt file showing the footprints infomation, and a hbond.txt file showing the hydrogen bond information. The footprint.txt file contains per residue interaction of the receptor with the ligand, for both reference molecule and the docked molecule. Shown below is an example of VDW footprints comparison. | ||
| + | |||
| + | [[File:2012_Dock_Tutorial_Footprints_1.png|thumb|375px|center|2012_DOCK_Tutorial_Footprints_1]] | ||
| + | |||
| + | [[File:2012_Dock_Tutorial_Footprints_2.png|thumb|375px|center|2012_DOCK_Tutorial_Footprints_2]] | ||
| + | |||
| + | Actually the upper picture and the bottom picture are comparing the same molecule to a same reference molecule. The difference is that the reference molecule for the upper case is the original ligand.mol2 without energy minimization! Obviously, the reference molecule is not correct in this case. As you can see, several very disfavored VDW interactions were observed for the reference molecule. In contrast, the bottom picture shows the correct comparison. | ||
| + | |||
| + | ==IX. Frequently Encountered Problems== | ||
===Barbara=== | ===Barbara=== | ||
| + | '''How to Open Files''' | ||
| + | |||
| + | If you want to view a file in chimera, first transfer the file from "herbie" or "seawulf" to your laptop using WinSCP. Then open Chimera on your laptop to open the files. | ||
| + | |||
===Woo Suk=== | ===Woo Suk=== | ||
| Line 626: | Line 730: | ||
You can change the fourth parameter from 0.0 to other values such as 0.1, 0.01, or 0.001. Because what you want is just to distinguish the very close spheres, you don't need large numbers. | You can change the fourth parameter from 0.0 to other values such as 0.1, 0.01, or 0.001. Because what you want is just to distinguish the very close spheres, you don't need large numbers. | ||
| − | === | + | ===Longfei=== |
| + | <b>Installation of DOCK</b> | ||
| + | |||
| + | If you want to install DOCK on your own desktop or laptop, and you are a beginner of Unix/Linux, you might encounter some problems during installation. A good reference for the installation is the DOCK User Manual(http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm). The problem I encountered was: after I used gnu as configuration file to configure the Makefile, I successfully made the <b>config.h</b> file, but when I was trying to build the DOCK executables, an error message " g77 command not found " appeared. And it is quite inconvenient to install the g77 command. A way to solve this problem is to open the <b>config.h</b> file, and manually change the <b>g77</b> in that file to <b>gfortran</b>. | ||
| + | |||
| + | <b>Use the Correct Path of Your File</b> | ||
| + | |||
| + | One frequently encountered problem is using the wrong path of your file. For example, when you use <b>sphere_selector</b> to generate spheres around the ligand, it requires two files--one is the spheres file .sph and another is your ligand file, but your ligand file is probably not in your working directory! So you need either copy your ligand file to the current directory or specify the correct path of your ligand file like below: | ||
| + | |||
| + | sphere_selector 1DF8.receptor.sph ../01-dockprep/1DF8.ligand.mol2 10.0 | ||
| + | |||
| + | This is also important when you run DOCK. Remember to use the correct path and name of the your file. | ||
| + | |||
| + | <b>Notice the Shell That You are Using</b> | ||
| + | |||
| + | This tutorial is based on the shell <b>tcsh</b>, and maybe you are using a different shell. This will not cause any trouble in most of the time, but you may want to notice this in some circumstances, for example when you want to execute some <b>.csh</b> file. One way to solve this problem is that simply type "tcsh" in the command line, and you will be running tcsh in another shell. | ||
| + | |||
===Roman=== | ===Roman=== | ||
| + | One of the problems one may encounter with grid creation is the sampling location for ligand binding. It is important to create a grid and dock the ligand to the correct location in the protein. If the grid is created in the incorrect location, the ligand binding sites will be sampled within the grid, and the best binding site will not be sampled. In our case, we are docking it to an area where a ligand is known to bind to streptavidin. However, this may not always be the case. | ||
| + | |||
| + | One solution is to dock to the center of mass of the spheres clusters created with spheregen and assess the center of mass as viable or not based on your chemical intuition. | ||
| + | |||
| + | Another way is to assess the protein for buried sites where there may also be a number of spheres. Both are options in the showbox program. | ||
| + | |||
| + | Finally, one can attempt to find proteins with similar sequence and determine where their ligand binding sites are, and then pick the binding site in the protein you are docking to using that information. | ||
| + | |||
===Quan=== | ===Quan=== | ||
===Hui=== | ===Hui=== | ||
| + | '''Keep the previous dock6 output .mol2 files''' | ||
| + | |||
| + | When we run dock6 program, the output file would be "1DF8.output_scored.mol2", with the prefix "1DF8.output" that you specified in dock.in file. In the case that you would have several runs of dock6 program with the output file of the same path (in the same directory), and you want to keep the result for each run, you need to rename "1DF8.output_scored.mol2" file to another one, otherwise the old .mol2 file will be overwritten by the new .mol2 file generated in the next run. | ||
| + | |||
| + | For example, you can rename the old .mol2 file generated by the first run and then perform the second run. | ||
| + | |||
| + | $mv 1DF8.output_scored.mol2 first_1DF8.output_scored.mol2 | ||
| + | |||
===Tuoling=== | ===Tuoling=== | ||
| + | |||
| + | '''Some recommended parameters we can change when running dock''' | ||
| + | |||
| + | max_orientations 500 | ||
| + | min_anchor_size 40 | ||
| + | pruning_conformer_score_cutoff 25 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | |||
| + | Explanations of these parameters can be found on the website provided by Long Fei. | ||
| + | Increase the number of these parameters can lead to increased running time. But it enables more thorough cycling and may give you better results. | ||
| + | |||
===Yuchen=== | ===Yuchen=== | ||
| + | ====Always Remember Your Path==== | ||
| + | |||
| + | One situation often appears is that the program will return an error saying "Could not open 1DF8.ligands.mol2 for reading." or something like that indicating that the program couldn't find the input file you specified in you <b>dock.in</b> or <b>vs.in</b> or some other files. In this case, the most often reason is that the path you specified in your input file is wrong. So the first thing you might want to do is to check those paths, pay special attention to those paths where you use "<b>.</b>" to indicate the same folder and "<b>..</b>" to indicate the folder up one level because this is the place where you are most likely to make mistakes. If you are not sure of the usage of "<b>.</b>" and "<b>..</b>", it will be safer for you to use the absolute paths of the files. And you can use the command <b>pwd</b> to ask the system where you are if you don't know that. | ||
| + | |||
===Yunting=== | ===Yunting=== | ||
| + | ====Delete Spheres Manually==== | ||
| + | We need to play some tricks if we want to manually select the spheres in <b>selected_spheres.sph</b>. | ||
| + | |||
| + | 1. Transfer <b>sph</b> file to <b>pbd</b> file by using <b>showsphere</b>.<br> | ||
| + | 2. Open the <b>pdb</b> file in Chimera and choose the sphere we want to delete.<br> | ||
| + | 3. Identify the number of that sphere by <b>Actions->Label->residue->specifier</b>.<br> | ||
| + | 4. Open <b>sph</b> file, delete that sphere and modify the number of spheres in that cluster.<br> | ||
| + | DOCK spheres within 6.0 ang of ligands | ||
| + | cluster 1 number of spheres in cluster 22 | ||
| + | 60 28.86000 11.09173 4.97943 2.337 586 0 0 | ||
| + | 67 28.67840 9.74620 12.13940 1.400 60 0 0 | ||
| + | 161 27.11509 13.42709 13.48051 1.401 385 0 0 | ||
| + | 174 27.79940 12.82468 12.04078 2.475 480 0 0 | ||
| + | 183 34.13903 14.01393 7.31591 3.590 691 0 0 | ||
| + | 187 36.78641 17.02434 9.54436 3.481 691 0 0 | ||
| + | 385 30.01753 14.79655 14.46633 1.402 174 0 0 | ||
| + | 463 25.74631 14.83304 9.98248 1.400 589 0 0 | ||
| + | |||
===Kip=== | ===Kip=== | ||
| + | |||
| + | ====Molecule preparation for molecular footprinting==== | ||
| + | |||
| + | A sample script: | ||
| + | |||
| + | #!/bin/tcsh | ||
| + | |||
| + | #check programs | ||
| + | foreach prog (antechamber parmchk tleap) | ||
| + | set prog_version = `which $prog` | ||
| + | if ( $prog_version == /nfs/user03/tbalius/amber10_ompi/bin/$prog ) then | ||
| + | echo $prog OK, current path is: `which $prog` | ||
| + | else | ||
| + | echo please double-check $prog path, current path is: `which $prog` | ||
| + | endif | ||
| + | end | ||
| + | |||
| + | #set your home directory on seawulf | ||
| + | set mydir = "$HOME" | ||
| + | |||
| + | # set up your ligands | ||
| + | foreach lig (ligand_name1 ligand_name2 ligand_name3) | ||
| + | |||
| + | #set working dir and go there | ||
| + | echo setting up directory for: $lig | ||
| + | mkdir "$mydir/DOCK_project2/10-ligandprep-top2mol2/$lig" | ||
| + | set workdir = "$mydir/DOCK_project2/10-ligandprep-top2mol2/$lig" | ||
| + | cd $workdir | ||
| + | cp ../$lig.mol2 . | ||
| + | echo 'Done!' | ||
| + | |||
| + | #convert mol2 to pdb and prep files | ||
| + | echo converting mol2 to pdb and prep files | ||
| + | antechamber -i $lig.mol2 -fi mol2 -o $lig.ante.pdb -fo pdb | ||
| + | antechamber -i $lig.mol2 -fi mol2 -o $lig.ante.prep -fo prepi | ||
| + | echo 'Done!' | ||
| + | |||
| + | #check for missing parameters | ||
| + | echo checking parameters for ligand: $lig | ||
| + | parmchk -i $lig.ante.prep -f prepi -o $lig.ante.frcmod | ||
| + | echo 'Done!' | ||
| + | |||
| + | #kludgey way to make input files | ||
| + | echo making input file for ligand: $lig | ||
| + | echo "set default PBradii mbondi2" > tleap.$lig.in | ||
| + | echo "source leaprc.ff99SB" >> tleap.$lig.in | ||
| + | echo "loadoff ../rizzo_amber7.ionparms/ions.lib" >> tleap.$lig.in | ||
| + | echo "loadamberparams ../rizzo_amber7.ionparms/ions.frcmod" >> tleap.$lig.in | ||
| + | echo "source leaprc.gaff" >> tleap.$lig.in | ||
| + | echo "loadamberparams ../rizzo_amber7.ionparms/gaff.dat.rizzo" >> tleap.$lig.in | ||
| + | echo "loadamberparams $lig.ante.frcmod" >> tleap.$lig.in | ||
| + | echo "loadamberprep $lig.ante.prep" >> tleap.$lig.in | ||
| + | echo "lig = loadpdb $lig.ante.pdb" >> tleap.$lig.in | ||
| + | echo "saveamberparm lig $lig.gas.leap.prm7 $lig.gas.leap.rst7" >> tleap.$lig.in | ||
| + | echo "solvateBox lig TIP3PBOX 10.0" >> tleap.$lig.in | ||
| + | echo "saveamberparm lig $lig.wat.leap.prm7 $lig.wat.leap.rst7" >> tleap.$lig.in | ||
| + | echo "charge lig" >> tleap.$lig.in | ||
| + | echo "quit" >> tleap.$lig.in | ||
| + | echo 'Done!' | ||
| + | |||
| + | #run tleap | ||
| + | echo running tleap for ligand: $lig | ||
| + | tleap -s -f tleap.$lig.in > tleap.$lig.out | ||
| + | echo 'Done!' | ||
| + | |||
| + | #make a new mol2 file | ||
| + | echo creating final mol2 file for ligand: $lig | ||
| + | top2mol2 -p $lig.gas.leap.prm7 -c $lig.gas.leap.rst7 -o $lig.gas.leap.mol2 | ||
| + | echo 'Done!' | ||
| + | |||
| + | #make a new pdb file | ||
| + | echo creating final pdb file for ligand: $lig | ||
| + | ambpdb -p $lig.gas.leap.prm7 -tit "pdb" < $lig.gas.leap.rst7 > $lig.gas.leap.pdb | ||
| + | echo 'Done!' | ||
| + | end | ||
| + | |||
====Keeping your data in sync==== | ====Keeping your data in sync==== | ||
One problem you may encounter is that you want to run your jobs on Seawulf, but you also want the same files on a mathlab computer (or your home computer) for analysis. One option is to use RSYNC to keep your files on Seawulf and Herbie (or any mathlab computer) in sync. For example, if you want to move your DOCK Tutorial files to Seawulf, do the following from Herbie or any mathlab computer: | One problem you may encounter is that you want to run your jobs on Seawulf, but you also want the same files on a mathlab computer (or your home computer) for analysis. One option is to use RSYNC to keep your files on Seawulf and Herbie (or any mathlab computer) in sync. For example, if you want to move your DOCK Tutorial files to Seawulf, do the following from Herbie or any mathlab computer: | ||
| Line 654: | Line 899: | ||
If you make a mistake and need to delete a job from the queue, first list all your queued or running jobs: | If you make a mistake and need to delete a job from the queue, first list all your queued or running jobs: | ||
| − | qstat -u | + | qstat -u username |
*IMPORTANT: If you get no output from qstat, it means that whatever jobs you have submitted are done! | *IMPORTANT: If you get no output from qstat, it means that whatever jobs you have submitted are done! | ||
Latest revision as of 12:22, 24 October 2012
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Markup as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2012.
Contents
- 1 I. Introduction
- 2 II. Preparing the Receptor and Ligand
- 3 III. Generating Receptor Surface and Spheres
- 4 IV. Generating Box and Grid
- 5 V. Docking a Single Molecule for Pose Reproduction
- 6 VI. Virtual Screening
- 7 VII. Running DOCK in Serial and in Parallel on Seawulf
- 8 VIII. Molecular Footprints
- 9 IX. Frequently Encountered Problems
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
Streptavidin & Biotin
Streptavidin is a tetrameric prokaryote protein that binds the co-enzyme biotin with an extremely high affinity. The streptavidin monomer is composed of eight antiparallel beta-strands which folds to give a beta barrel tertiary structure. A biotin binding-site is located at one end of each β-barrel, which has a high affinity as well as a high avidity for biotin. Four identical streptavidin monomers associate to give streptavidin’s tetrameric quaternary structure. The biotin binding-site in each barrel consists of residues from the interior of the barrel, together with a conserved Trp120 from neighboring subunit. In this way, each subunit contributes to the binding site on the neighboring subunit, and so the tetramer can also be considered a dimer of functional dimers.
Biotin is a water soluble B-vitamin complex which is composed of an ureido (tetrahydroimidizalone) ring fused with a tetrahydrothiophene ring. It is a co-enzyme that is required in the metabolism of fatty acids and leucine. It is also involved in gluconeogenesis.
Organizing Directories
While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify. For this tutorial, we will use something similar to the following:
~username/AMS536/DOCK-Tutorial/00-original-files/
/01-dockprep/
/02-surface-spheres/
/03-box-grid/
/04-dock/
/05-virtual-screen/
In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '1DF8'. The following sections in this tutorial will adhere to this directory structure / naming scheme.
II. Preparing the Receptor and Ligand
Downloading the PDB Structure (1DF8)
The Protein Data Bank contains atomic coordinates for more than 79,000 molecules and is accessible via the internet from PDB. The target (protein or nucleic acid) is accessible by PDB ID, molecular name, or author. After entering the PDB ID, molecule name, or author, click Download Files, then select PDB File (text). A window will open; click on Save File → Downloads.
Preparing the Ligand and Enzyme in Chimera
If you are on "herbie" you can access Chimera directly by typing in Chimera at the prompt. Otherwise, download Chimera to your desktop and obtain your protein/nucleic acid complex of interest using Fetch (by ID).
Open Your Protein of Interest and Delete Unwanted Molecules:
Click on Open under the File menu and find where you saved your pdb file.
You only need one of the monomers to perform docking. Remove chain B by carrying out the following:
- Select → Chain → B → All.
- Actions →Atoms → Delete.
To delete water molecules and/or other ligands, go to Tools → Structure Edit → Dock Prep. Check all boxes and click okay. This will walk you through the steps needed to prepare the complex for docking, and will also assign partial charges to the protein and the ligand. Choose am 1-bcc for charges.
Save the file as 1DF8.dockprep.mol2.
This file contains a conformation of the complex with hydrogen atoms. The grid calculation is based on the receptor with its hydrogen atoms. The grid score is an energy calculation that is based on the following equation:
E GRID = E VDW + E ES. The score is an approximation of the molecular mechanics' energy function and it considers only through space interactions.
Create a Receptor File:
Go to Select → Residue → BTN. Then go to Actions → Atoms → Delete.
Save the file as 1DF8.receptor.mol2.
Create a Receptor File with No Hydrogen Atoms:
Go to Select → Chemistry → Element → H → Delete.
Save a PDB file as 1DF8.receptor.noH.pdb.
Create a Ligand File:
Open only the 1DF8.dockprep.mol2.
Go to Select → Chain → Delete. This will allow you to have only the ligand.
Save the file as 1DF8.ligand.mol2.
These are the files that you will need to continue with rigid or flexible docking.
III. Generating Receptor Surface and Spheres
Receptor Surface
To generate an enzyme surface, first open the receptor pdb file with the hydrogen atoms removed (1DF8.receptor.noH.pdb). Next, go to Actions -> Surface -> Show. Note that for DOCK calculation hydrogen atoms are considered, but for generating enzyme surface and spheres, it is necessary to use the protein without hydrogens.
Recent versions of Chimera include a Write DMS tool that facilitates calculation of the molecular surface. Go to Tools -> Structure Editing -> Write DMS. Save the surface as 1DF8.receptor.dms.
The Write DMS tool will "roll" a small probe (default radius = 1.4 Angstroms = Size of a water molecule) over the surface of the enzyme and calculate the surface normal at each point. DMS (distributed molecular surface) file is subsequently used as input file for sphgen.
Spheres
To generate docking spheres, we need to use a command line program called sphgen. To run the sphgen, we need a input file named INSPH.
1DF8.receptor.dms #molecular surface file that we got from previous step R #sphere outside of surface (R) or inside surface (L) X #specifies subset of surface points to be used (X=all points) 0.0 #prevents generation of large spheres with close surface contacts(defalut=0.0) 4.0 #maximum sphere radius in angstroms (default=4.0) 1.4 #minimum sphere radius in angstroms (default=radius of probe) 1DF8.receptor.sph #clustered spheres file that we want to generate
Save the INSPH file. Then use this command to generate the spheres file:
sphgen -i INSPH -o OUTSPH
-i means input; -o means output.
You should get two output files: OUTSPH and 1DF8.receptor.sph. The OUTSPH file should similar to this:
density type = X reading 1DF8.receptor.dms type R # of atoms = 881 # of surf pts = 10771 finding spheres for 1DF8.receptor.dms dotlim = 0.000 radmax = 4.000 Minimum radius of acceptable spheres? 1.4000000 output to 1DF8.receptor.sph clustering is complete 28 clusters
You can also open the spheres file that we generated in this step (1DF8_receptor.sph). This file contains detailed information about the spheres, which are divided into 28 clusters. Cluster 0 in the end of the spheres file is a combination of all the clusters.
In order to visualize the generated spheres, you can use a program called showsphere. Showsphere is an interactive program. In the command line, simply type
showsphere
You will be asked the following questions:
Enter name of sphere cluster file: 1DF8.receptor.sph Enter cluster number to process (<0 = all): -1 Generate surfaces as well as pdb files (<N>/Y)? N Enter name for output file prefix: 1DF8.output. Process cluster 0 (contains ALL spheres) (<N>/Y)? N
In this case, 28 pdb files should be generated.
An alternative way to do this is to create an input file, showsphere.in, as follows:
1DF8.receptor.sph -1 N 1DF8.output N
Then type the command showsphere
showshpere < showsphere.in
After generating the pdb files by showsphere, you can visualize each cluster by UCSF Chimera. For example, open 1DF8.output_1.pdb by Chimera, then choose Actions->Atoms/Bonds->sphere, you will be able to see the spheres in cluster 1. You can also open the receptor file (1DF8.receptor.mol2) at the same time, then choose Presets->Interactive 3(hydrophobicity surface), then again choose Actions->Atoms/Bonds->sphere, you will be able to see what the spheres in cluster 1 look like on the enzyme surface.
There are over 500 spheres in the spheres file(1DF8.receptor.sph). However, we're only interested in docking the ligand into the active site. Therefore we need to select only those spheres which are inside the active site, using sphere_selector program.
sphere_selector 1DF8.receptor.sph ../01-dockprep/1DF8.ligand.mol2 10.0
Sphere_selector filters the output from sphgen, selecting all spheres within a user-defined radius of a target molecule. In this case, we selected the spheres within 10 angstroms of our ligand. A file called selected_spheres.sph should be created, showing the spheres that are selected. You can again visualize it by showsphere. You can also change the radius (to 8 angstroms or 6 angstroms) or manually edit the file selected_spheres.sph so that you can select the spheres you want. In this tutorial, spheres within 6 angstroms of the ligand are used for the next step (with one sphere which does not belong to the active site being manually deleted).
IV. Generating Box and Grid
Box
In order to speed up docking calculations, DOCK generates a fine grid. At each point in the grid, electrostatic and the VDW probes' energies are precomputed. The energies are computed using a molecular force field. To determine the dimensions of the grid, however, we first generate a box that contains the outer boundaries for the grid calculation. The dimensions and location of the box can be determined using a program called showbox.
First create a directory where you will place the grid files.
$mkdir 03-box-grid $cd 03-box-grid
showbox can be used interactively or a file with predetermined answers can be fed into the program.
The program asks the questions depicted in the diagram the right:
To run the program in the interactive mode, run
$showbox
To feed the answers to the questions, run
$showbox < showbox.in
For example, showbox.in can contain:
Y 5 ../02-surface-spheres/selected_spheres.sph 1 1DF8.box.pdb
Y means we use automatic box construction, 5 is the extra margin to be enclosed around our ligand (in Angstroms), selected_spheres.sph is the sphere file we generated, 1 corresponds to the cluster number in the selected_spheres.sph file, and 1DF8.box.pdb is the output file. We can open the output box file in chimera to make sure the box is in the right place.
Grid
Now let's generate a grid within our box. We will use the energy scoring method to generate a grid, resulting in three additional files with extensions *.nrg, *.bmp, and *.out. The *.nrg file contains the energy scoring, *.bmp contains the size, position and grid spacing and determines whether there are any overlaps with receptor atoms.
To generate the grid we will use the grid program. This program can either be used interactively, or an input file can be fed in, just like the showbox program.
Usage: grid [-i [input_file]] [-o [output_file]] ...
[-standard_i/o] [-terse] [-verbose]
-i: read from grid.in or input_file, standard_in otherwise
-o: write to grid.out or output_file (-i required),
standard_out otherwise
-s: read from and write to standard streams (-i and/or -o illegal)
-t: terse program output
-v: verbose program output
For our grid.in file, we will use the following answers:
compute_grids yes grid_spacing 0.3 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../01-dockprep/1DF8.receptor.mol2 box_file 1DF8.box.pdb vdw_definition_file /opt/software/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Line by line:
- compute scoring grids (yes)
- what is the distance between grid points along each axis (in Angstroms).
- write up coordinates of the receptor into a new file
- compute contact grid? default is no
- compute energy score? yes - we are using this method to compute force fields on probes
- the max distance between atoms for the energy contribution to be computed
- atom_model u means united atom model where atoms are attached to hydrogens, and a stands for all-atom model, where hydrogens on carbons are treated separately
- attractive component stands for exponent of the attractive LJ term in VDW potential
- repulsive component stands for exponent in the repulsive LJ term in VDW potential
- distance dielectric stands for the dielectric constant to be linearly dependent on distance
- distance dielectric factor is the coefficient of the dielectric
- bump filter flag determines if we want to screen orientation for clashes before scoring and minimization
- bump_overlap stands for the fraction of allowed overlap where 1 corresponds to no allowed overlap and 0 corresponds to full overlap being permitted.
- our receptor file
- the box file we generated in the Box section
- VDW parameters file
- Prefix for the grid file name. All the extensions will be generated automatically.
V. Docking a Single Molecule for Pose Reproduction
Docking and Results
Change directory into “04-dock” and create an empty input file called dock.in
$touch dock.in #create a file called “dock.in”
Run dock6.4
$dock6 -i dock.in
Alternatively:
$dock6 -i dock -v # “-v” option allows you to print out the information regarding each growth step on terminal
Or:
$dock6 -i dock -o dock.bf.out -v # “-o” allows you to write the aforementioned information into an output file named “dock.bf.out”
1st run:
Notice that running dock6 with an empty input file will require you to answer a series of questions. For the first run we will deactivate most of the features by selecting “no”. Parameters requiring specification are listed below:
ligand_atom_file [database.mol2] (): ../01-dockprep/1DF8.ligand.mol2 ligand_outfile_prefix [output] (): 1DF8.output
What the program is doing in the 1st run is to take the ligand.mol2 file and directly generate an output file without any additional manipulations. You would expect it to be exactly the same as the original pose. You can open it in Chimera by using the “ViewDock” function under “Tools->Surface/Binding Analysis”. We will not show the result here.
2nd run:
The real experiment begins here. Notice that we selected “no” for most of the functions. This time we will try change some parameters in the dock.in file.
$vim dock.in
Parameters being changed are listed below:
orient_ligand yes
The "orient_ligand" option tells the program whether to try orientations different from the pose in the original .mol2 file. Note that because of the change, additional questions are asked by the program. For simplicity we keep most of the answers as default. File paths that need to be specified are listed below:
receptor_site_file [receptor.sph] ():../02-surface-spheres/selected_spheres_06A.sph vdw_defn_file [vdw.defn] ():../03-grid/vdw_AMBER_parm99.defn flex_defn_file [flex.defn] ():../03-grid/flex.defn flex_drive_file [flex_drive.tbl] ():../03-grid/flex_drive.tbl
The receptor_site_file should be the selected spheres file (.sph) generated in a previous step (02 surface and spheres), according to which the program orients the ligand.
The vdw_defn_file instructs the dock6 program to use the Van der Waals parameter sets from the AMBER force field.
The flex_defn_file and the flex_drive_file contain the information required by the program to sample conformations.
The result is shown below. As you may notice the result doesn't look very good.
3rd Run:
Here we will further specify parameters to improve the result.
calculate_rmsd yes score_molecules yes num_scored_conformers 10
Again, further questions will be asked when you run the program. Keep most answers as the default values. Paths requiring specification are listed below:
grid_score_grid_prefix [grid] ():../03-grid/1DF8.grid
Note that the above specification will tell the program to load the 1DF8.grid.nrg file you generated in the previous step (grid) for scoring.
simplex_max_iterations [1000] ():20 write_conformations [yes] (yes no):no cluster_rmsd_threshold [2.0] ():0.2
The simplex_max_iteration parameter specifies number of minimization cycles.
Note the program will cluster poses that are very close together (rmsd smaller than the threshold specified in the cluster_rmsd_threshold parameter) into a cluster.
This time the program returned the best 10 poses. The one with the best grid score (-52.8, shown blue) superimposed quite well with the original pose in the crystal structure (RMSD = 0.71). You can view the grid scores in ViewDock by selecting “Column->Show->Grid Score”
4th Run:
This time we will set the ligand as flexible:
flexible_ligand yes
Further questions will be asked during the run. Keep most of the answers as the default values except for:
simplex_grow_max_iterations [20] ():500
The simplex_grow_max_iteration specifies the maximum number of iterations per cycle per growth step.
Notice that the run is significantly slower this time. Again the best pose generated (-64.8) is shown in blue. The grid score improved significantly but the RMSD did not change much (0.79)
5th Run:
This time we will turn the bump filter on:
bump_filter yes
The bump_filter option filters out conformations that cause clashes between atoms.
Further questions will be asked during the run. Keep answers as the default values. Specify the following path:
bump_grid_prefix [grid] ():../03-grid/1DF8.grid
Note that the path tells the program to access the .bmp file generated in the previous grid step.
The best pose (grid score -65.3) is again shown in blue. Notice that turning on the bump filter does not alter either the grid score or the RMSD (0.78).
Remember you can tweak any parameter in the dock.in file instead of keeping them as default.
Following is the final dock.in file used during the fifth run:
ligand_atom_file ../01-dockprep/1DF8.ligand.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd yes
use_rmsd_reference_mol no
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file ../02-surface-spheres/selected_spheres_06A.sph
max_orientations 500
critical_points no
chemical_matching no
use_ligand_spheres no
use_internal_energy yes
internal_energy_rep_exp 12
flexible_ligand yes
min_anchor_size 40
pruning_use_clustering yes
pruning_max_orients 100
pruning_clustering_cutoff 100
pruning_conformer_score_cutoff 25
use_clash_overlap no
write_growth_tree no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../03-grid/1DF8.grid
dock3.5_score_secondary no
continuous_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
amber_score_secondary no
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_anchor_max_iterations 500
simplex_grow_max_iterations 500
simplex_grow_tors_premin_iterations 0
simplex_random_seed 0
simplex_restraint_min no
atom_model all
vdw_defn_file ../03-grid/vdw_AMBER_parm99.defn
flex_defn_file ../03-grid/flex.defn
flex_drive_file ../03-grid/flex_drive.tbl
ligand_outfile_prefix 1DF8.output
write_orientations no
num_scored_conformers 10
write_conformations no
cluster_conformations yes
cluster_rmsd_threshold 0.2
rank_ligands no
Now we will write up a script for submitting your dock job to Seawulf. Create a script called “sub.dock.csh”
$vim sub.dock.csh
wherein you write:
#! /bin/tcsh #PBS -l nodes=1:ppn=1 #PBS -l walltime=01:00:00 #PBS -o zzz.qsub.out #PBS -j oe cd ~/AMS536/DOCK_tutorial/05-dock_qsub /nfs/user03/sudipto/dock6/bin/dock6 -i dock.in -o dock.out
This will request 1 processor from the cluster for your job. When you are submitting the job:
$qsub sub.dock.csh
Note that you might have to make the script executable before running it:
$chmod +x sub.dock.csh
Results
VI. Virtual Screening
Virtual Screening Preparation
Virtual screening is a widely used method in computational drug design. It searches large libraries of chemical compounds to identify favorable structures that bind to a target molecule. To perform virtual screening, we use ligands.mol2, a mol2 file which contains 101 small molecules to be the virtual library. The computational cost is reasonable for a quick search. Usually we use larger molecule set from chemical database, such as ZINC(http://zinc.docking.org/).
Since the computational cost of virtual screening is much higher than single-ligand docking, it is better for us to run it on Seawulf. We need to compress the whole DOCK-Tutorial directory, copy it to Seawulf and compress it.
tar -cvf DOCK-Tutorial.tar DOCK-Tutorial/ scp DOCK-Tutorial sw:/nfs/user03/usrname/AMS536 tar -xvf DOCK-Tutorial.tar DOCK-Tutorial
Virtual Screening Protocol
The purpose of virtual screening is different from single molecule docking, so we need to modify our previous docking script dock.in to vs.in. We can see the difference between the two files.
1c1 < ligand_atom_file ligands.mol2 --- > ligand_atom_file ../01-dockprep/1DF8.ligand.mol2 56,59c56,59 < vdw_defn_file /nfs/user03/sudipto/dock6/parameters/vdw_AMBER_parm99.defn < flex_defn_file /nfs/user03/sudipto/dock6/parameters/flex.defn < flex_drive_file /nfs/user03/sudipto/dock6/parameters/flex_drive.tbl < ligand_outfile_prefix 1DF8.vs.output --- > vdw_defn_file ../03-box-grid/vdw_AMBER_parm99.defn > flex_defn_file ../03-box-grid/flex.defn > flex_drive_file ../03-box-grid/flex_drive.tbl > ligand_outfile_prefix 1DF8.output 61c61 < num_scored_conformers 1 --- > num_scored_conformers 10 63c63,64 < cluster_conformations no --- > cluster_conformations yes > cluster_rmsd_threshold 0.2
num_scored_conformers: 1 -> 10 In virtual screening, we only need the most favorable pose of each candidate molecule and compare them.
cluster_conformations: yes -> no Slightly different conformations are not clustered together. They are treated as different conformations in the docking process.
In order to generate different search spaces, we can modify some other parameters.
max_orientations: The maximal number of anchor orientations that will be generated.
min_anchor_size: The minimum number of atoms for an anchor.
pruning_use_clustering: Pruning conformations during the clustering process.
use_internal_energy: Using repulsive VDM to avoid internal clashes.
Parallel computing can reduce the running time of virtual screening. Here is our job submission script sub.virtual_screen.csh.
#! /bin/tcsh
#PBS -l nodes=4:ppn=2
#PBS -l walltime=01:00:00
#PBS -o zzz.qsub.out
#PBS -j oe
#PBS -V
set nprocs = `wc -l $PBS_NODEFILE | awk '{print $1}'`
echo "Running on ${nprocs} processors"
echo ""
echo "processor list are:"
cat $PBS_NODEFILE
cd ~/AMS536/DOCK_Tutorial/06-virtualscreen
mpirun -np $nprocs dock6.mpi -i vs.in -o vs.out
Finally, we can submit the job and perform virtual screening.
qsub sub.virtual_screen.csh
Virtual Screening Results
After performing virtual screening on Seawulf or any other computer, if you are using a parallel computer, you should get a multi-mol2 file 1DF8.vs.output_scored.mol2 which contains the mol2 files of all succesfully docked ligands, a vs.out file which contains the dock results of successfully docked ligands, and vs.out.1 through vs.out.7 which contain dock results from different nodes you use (the number will vary according to the number of nodes you use, here we use 8 nodes as mentioned before).
The vs.out file is returned by the leading node and contains the information of each succesfully docked ligands, looks like this:
Molecule: ZINC33171556
Anchors: 1
Orientations: 500
Conformations: 116
Grid Score: -52.373383
Grid_vdw: -51.643253
Grid_es: -0.730129
Int_energy: 6.125062
The vs.out.1 through vs.out.2 files are returned by other nodes processing those ligands, separately. For those succesfully docked ligands, the file will return the elapsed time for docking, and for those not succesfully docked, it will return an error massege like this:
Molecule: ZINC20605433
Elapsed time: 0 seconds
ERROR: Could not complete growth.
Confirm that the grid box is large enough to contain the ligand,
and try increasing max_orientations.
If you download the multi-mol2 file from seawulf using:
scp sw:~/AMS536/DOCK_tutorial/06-virtual-screen/1DF8.vs.output_scored.mol2 ./
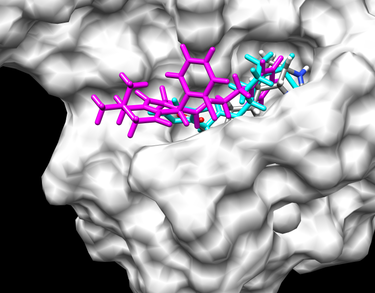
Now you have this multi-mol2 file 1DF8.vs.output_scored.mol2 on your local machine, you can actually open this file in Chimera to visually check you docking results and do some visual analysis. But it's not a good idea to directly open your multi-mol2 file because it contains information of all 47 succesfully docked ligands, if you just open this file, it will be pretty messy. So what you will do is, first you open the receptor file 1DF8.receptor.mol2 and the ligand 1DF8.ligands.mol2 as a reference. And then use the ViewDock function of Chimera to look at your 47 ligands one or however many you want at a time. You can find ViewDock via Tools -> Surface/Binding Analysis -> ViewDock, and then find your 1DF8.vs.output_scored.mol2 file in your own directory and click on open. Now a new window of ViewDock will pop out and there will be an extra ligands in Chimera main window which is the highlighted ligand in ViewDock window. You can look at the ligands one at a time, you can also hold ctrl and click on different ligands to view them at the same time, this will give you a direct idea of how good these ligands dock. The other thing you can do is, the multi-mol2 file 1DF8.vs.output_scored.mol2 contains the energy score of every ligand, so in ViewDock window, you can go to Column -> show -> Grid Score to show the grid score, and then you can click on the head of the column to rank order all the ligands by their grid scores.The picture showing here is the best and worst scored ligands of out calculation, the best scroed one in cyan and the worst scored on in magenta, and the reference ligand is colored according to elements. As you can see, the best scored ligand fits in the binding pocket very well, but the worst scored one almost sticks out of the binding pocket.
VII. Running DOCK in Serial and in Parallel on Seawulf
The Seawulf Cluster has 235 dual processor nodes (2 processors per node), for a total of 470 individual processors. These are 3.4Ghz Intel Pentium IV Xeon processors from Dell. Each node has 2GB attached RAM and a 40GB hard disk.
Typically you will use one processor on a single node to dock one ligand. If you are docking multiple ligands, you can use more than processor in parallel mode, but you should never use more processors than you have ligands.
Running DOCK in Serial on a Single Processor
The following sample code can be used to run DOCK on one processor on a single node:
#!/bin/tcsh #PBS -l nodes=1:ppn=1 #PBS -l walltime=01:00:00 #PBS -N dock6 #PBS -M user@ic.sunysb.edu #PBS -j oe #PBS -o pbs.out cd /nfs/user03/sudipto/DOCK_Tutorial /nfs/user03/sudipto/dock6/bin/dock6 -i dock.in -o dock.out
Here is an explanation of the code and format:
#!/bin/tcsh #Execute script with tcsh #PBS -l nodes=1:ppn=1 #Use one node, and one processor per node, so one single processor #PBS -l walltime=01:00:00 #Allow 1 hour for your job run #PBS -N dock6 #Name of your job #PBS -M user@ic.sunysb.edu #Get an email notifying you when your job is completed #PBS -j oe #Combine the output and error streams into a single output file #PBS -o pbs.out #Name of your output file cd /path-to-your-home-directory-on-seawulf/DOCK_Tutorial #Change to your home directory and folder with dock files /nfs/user03/sudipto/dock6/bin/dock6 -i dock.in -o dock.out #Specifies path to dock executable and provide input and output filenames
Running DOCK in Parallel using MPI
The following sample code can be used to run DOCK on 4 nodes, using both processors on each node, for a total of 8 processors.
#! /bin/tcsh
#PBS -l nodes=4:ppn=2
#PBS -l walltime=24:00:00
#PBS -o zzz.qsub.out
#PBS -j oe
#PBS -V
set nprocs = `wc -l $PBS_NODEFILE | awk '{print $1}'`
echo "Running on ${nprocs} processors"
cd /nfs/user03/sudipto/DOCK_Tutorial
cp /nfs/user03/sudipto/dock6/parameters/vdw_AMBER_parm99.defn .
cp /nfs/user03/sudipto/dock6/parameters/flex* .
mpirun -np $nprocs /nfs/user03/sudipto/dock6/bin/dock6.mpi -i dock.in -o dock.out
For more information, see PBS Queue
Serial Calculation for Pose Reproduction
Parallel Virtual Screen
VIII. Molecular Footprints
Preparation (minimization) of Reference Molecule
The footprint of the reference molecule guides the selection of molecules/poses in the footprint rescoring method, therefore careful preparation of this reference molecule is necessary. Typically the reference molecule will be an endogenous substrate or known inhibitor from an x-ray crystallographic structure.
Preferably, the molecule should be prepared with hydrogens and charges using AMBER and subsequently converted to mol2 format using the 'top2mol2' command. This will provide you with a 'monotonic' mol2 file -- meaning that all the hydrogens will be listed with the corresponding protein residue, rather than separately at the end of the file (like Chimera's 'DockPrep' tool does).
An alternative way to generate the "monotonic" receptor file is to open the receptor.mol2 file with Chimera, save as pdb file, reopen the pdb file and go through DockPrep, and again save as a mol2 file. In this case, the new mol2 file will be monotonic.
Once the molecule is prepared, it should be minimized in the receptor using a tethered minimization. This is especially important if the crystal structure was refined without hydrogens (typically hydrogens are not used in refinement unless the resolution is ~1 Angstrom or better). Minimization is needed to help alleviate any steric clashes involving the newly added hydrogens (which would otherwise disrupt your footprint results).
An example input file for use on seawulf (using dock6.5):
ligand_atom_file ligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol no use_database_filter no orient_ligand no use_internal_energy yes internal_energy_rep_exp 12 flexible_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary yes continuous_score_secondary no cont_score_rec_filename receptor.mol2 cont_score_att_exp 6 cont_score_rep_exp 12 cont_score_rep_rad_scale 1 cont_score_use_dist_dep_dielectric yes cont_score_dielectric 4.0 cont_score_vdw_scale 1 cont_score_es_scale 1 descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/vdw_AMBER_parm99.defn flex_defn_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/flex.defn flex_drive_file /nfs/user03/tbalius/zzz.programs/dock6.5/parameters/flex_drive.tbl ligand_outfile_prefix xtalmin write_orientations no num_scored_conformers 1 rank_ligands no
Molecular footprints comparisons
After rescoring of the molecules by footprint-based score, typically three output files will be generated: a mol2 file showing the ligands information, a footprint.txt file showing the footprints infomation, and a hbond.txt file showing the hydrogen bond information. The footprint.txt file contains per residue interaction of the receptor with the ligand, for both reference molecule and the docked molecule. Shown below is an example of VDW footprints comparison.
Actually the upper picture and the bottom picture are comparing the same molecule to a same reference molecule. The difference is that the reference molecule for the upper case is the original ligand.mol2 without energy minimization! Obviously, the reference molecule is not correct in this case. As you can see, several very disfavored VDW interactions were observed for the reference molecule. In contrast, the bottom picture shows the correct comparison.
IX. Frequently Encountered Problems
Barbara
How to Open Files
If you want to view a file in chimera, first transfer the file from "herbie" or "seawulf" to your laptop using WinSCP. Then open Chimera on your laptop to open the files.
Woo Suk
Distinguishing overlapped spheres
Sometimes your 1DF8.output_1.pdb can have some overlapped spheres. In this case, you cannot find them before you delete one sphere and you have to repeat this one more time to delete another one.
In order to avoid the situation, you can adjust a parameter in INSPH file like following:
1DF8.receptor.dms R X 0.001 4.0 1.4 1DF8.receptor.sph
You can change the fourth parameter from 0.0 to other values such as 0.1, 0.01, or 0.001. Because what you want is just to distinguish the very close spheres, you don't need large numbers.
Longfei
Installation of DOCK
If you want to install DOCK on your own desktop or laptop, and you are a beginner of Unix/Linux, you might encounter some problems during installation. A good reference for the installation is the DOCK User Manual(http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm). The problem I encountered was: after I used gnu as configuration file to configure the Makefile, I successfully made the config.h file, but when I was trying to build the DOCK executables, an error message " g77 command not found " appeared. And it is quite inconvenient to install the g77 command. A way to solve this problem is to open the config.h file, and manually change the g77 in that file to gfortran.
Use the Correct Path of Your File
One frequently encountered problem is using the wrong path of your file. For example, when you use sphere_selector to generate spheres around the ligand, it requires two files--one is the spheres file .sph and another is your ligand file, but your ligand file is probably not in your working directory! So you need either copy your ligand file to the current directory or specify the correct path of your ligand file like below:
sphere_selector 1DF8.receptor.sph ../01-dockprep/1DF8.ligand.mol2 10.0
This is also important when you run DOCK. Remember to use the correct path and name of the your file.
Notice the Shell That You are Using
This tutorial is based on the shell tcsh, and maybe you are using a different shell. This will not cause any trouble in most of the time, but you may want to notice this in some circumstances, for example when you want to execute some .csh file. One way to solve this problem is that simply type "tcsh" in the command line, and you will be running tcsh in another shell.
Roman
One of the problems one may encounter with grid creation is the sampling location for ligand binding. It is important to create a grid and dock the ligand to the correct location in the protein. If the grid is created in the incorrect location, the ligand binding sites will be sampled within the grid, and the best binding site will not be sampled. In our case, we are docking it to an area where a ligand is known to bind to streptavidin. However, this may not always be the case.
One solution is to dock to the center of mass of the spheres clusters created with spheregen and assess the center of mass as viable or not based on your chemical intuition.
Another way is to assess the protein for buried sites where there may also be a number of spheres. Both are options in the showbox program.
Finally, one can attempt to find proteins with similar sequence and determine where their ligand binding sites are, and then pick the binding site in the protein you are docking to using that information.
Quan
Hui
Keep the previous dock6 output .mol2 files
When we run dock6 program, the output file would be "1DF8.output_scored.mol2", with the prefix "1DF8.output" that you specified in dock.in file. In the case that you would have several runs of dock6 program with the output file of the same path (in the same directory), and you want to keep the result for each run, you need to rename "1DF8.output_scored.mol2" file to another one, otherwise the old .mol2 file will be overwritten by the new .mol2 file generated in the next run.
For example, you can rename the old .mol2 file generated by the first run and then perform the second run.
$mv 1DF8.output_scored.mol2 first_1DF8.output_scored.mol2
Tuoling
Some recommended parameters we can change when running dock
max_orientations 500 min_anchor_size 40 pruning_conformer_score_cutoff 25 pruning_clustering_cutoff 100
Explanations of these parameters can be found on the website provided by Long Fei. Increase the number of these parameters can lead to increased running time. But it enables more thorough cycling and may give you better results.
Yuchen
Always Remember Your Path
One situation often appears is that the program will return an error saying "Could not open 1DF8.ligands.mol2 for reading." or something like that indicating that the program couldn't find the input file you specified in you dock.in or vs.in or some other files. In this case, the most often reason is that the path you specified in your input file is wrong. So the first thing you might want to do is to check those paths, pay special attention to those paths where you use "." to indicate the same folder and ".." to indicate the folder up one level because this is the place where you are most likely to make mistakes. If you are not sure of the usage of "." and "..", it will be safer for you to use the absolute paths of the files. And you can use the command pwd to ask the system where you are if you don't know that.
Yunting
Delete Spheres Manually
We need to play some tricks if we want to manually select the spheres in selected_spheres.sph.
1. Transfer sph file to pbd file by using showsphere.
2. Open the pdb file in Chimera and choose the sphere we want to delete.
3. Identify the number of that sphere by Actions->Label->residue->specifier.
4. Open sph file, delete that sphere and modify the number of spheres in that cluster.
DOCK spheres within 6.0 ang of ligands cluster 1 number of spheres in cluster 22 60 28.86000 11.09173 4.97943 2.337 586 0 0 67 28.67840 9.74620 12.13940 1.400 60 0 0 161 27.11509 13.42709 13.48051 1.401 385 0 0 174 27.79940 12.82468 12.04078 2.475 480 0 0 183 34.13903 14.01393 7.31591 3.590 691 0 0 187 36.78641 17.02434 9.54436 3.481 691 0 0 385 30.01753 14.79655 14.46633 1.402 174 0 0 463 25.74631 14.83304 9.98248 1.400 589 0 0
Kip
Molecule preparation for molecular footprinting
A sample script:
#!/bin/tcsh
#check programs
foreach prog (antechamber parmchk tleap)
set prog_version = `which $prog`
if ( $prog_version == /nfs/user03/tbalius/amber10_ompi/bin/$prog ) then
echo $prog OK, current path is: `which $prog`
else
echo please double-check $prog path, current path is: `which $prog`
endif
end
#set your home directory on seawulf
set mydir = "$HOME"
# set up your ligands
foreach lig (ligand_name1 ligand_name2 ligand_name3)
#set working dir and go there
echo setting up directory for: $lig
mkdir "$mydir/DOCK_project2/10-ligandprep-top2mol2/$lig"
set workdir = "$mydir/DOCK_project2/10-ligandprep-top2mol2/$lig"
cd $workdir
cp ../$lig.mol2 .
echo 'Done!'
#convert mol2 to pdb and prep files
echo converting mol2 to pdb and prep files
antechamber -i $lig.mol2 -fi mol2 -o $lig.ante.pdb -fo pdb
antechamber -i $lig.mol2 -fi mol2 -o $lig.ante.prep -fo prepi
echo 'Done!'
#check for missing parameters
echo checking parameters for ligand: $lig
parmchk -i $lig.ante.prep -f prepi -o $lig.ante.frcmod
echo 'Done!'
#kludgey way to make input files
echo making input file for ligand: $lig
echo "set default PBradii mbondi2" > tleap.$lig.in
echo "source leaprc.ff99SB" >> tleap.$lig.in
echo "loadoff ../rizzo_amber7.ionparms/ions.lib" >> tleap.$lig.in
echo "loadamberparams ../rizzo_amber7.ionparms/ions.frcmod" >> tleap.$lig.in
echo "source leaprc.gaff" >> tleap.$lig.in
echo "loadamberparams ../rizzo_amber7.ionparms/gaff.dat.rizzo" >> tleap.$lig.in
echo "loadamberparams $lig.ante.frcmod" >> tleap.$lig.in
echo "loadamberprep $lig.ante.prep" >> tleap.$lig.in
echo "lig = loadpdb $lig.ante.pdb" >> tleap.$lig.in
echo "saveamberparm lig $lig.gas.leap.prm7 $lig.gas.leap.rst7" >> tleap.$lig.in
echo "solvateBox lig TIP3PBOX 10.0" >> tleap.$lig.in
echo "saveamberparm lig $lig.wat.leap.prm7 $lig.wat.leap.rst7" >> tleap.$lig.in
echo "charge lig" >> tleap.$lig.in
echo "quit" >> tleap.$lig.in
echo 'Done!'
#run tleap
echo running tleap for ligand: $lig
tleap -s -f tleap.$lig.in > tleap.$lig.out
echo 'Done!'
#make a new mol2 file
echo creating final mol2 file for ligand: $lig
top2mol2 -p $lig.gas.leap.prm7 -c $lig.gas.leap.rst7 -o $lig.gas.leap.mol2
echo 'Done!'
#make a new pdb file
echo creating final pdb file for ligand: $lig
ambpdb -p $lig.gas.leap.prm7 -tit "pdb" < $lig.gas.leap.rst7 > $lig.gas.leap.pdb
echo 'Done!'
end
Keeping your data in sync
One problem you may encounter is that you want to run your jobs on Seawulf, but you also want the same files on a mathlab computer (or your home computer) for analysis. One option is to use RSYNC to keep your files on Seawulf and Herbie (or any mathlab computer) in sync. For example, if you want to move your DOCK Tutorial files to Seawulf, do the following from Herbie or any mathlab computer:
rsync -arv /home/username/DOCK_Tutorial/ sw:/nfs/user03/username/DOCK_Tutorial
- Note: The trailing slash on /home/username/DOCK_Tutorial/ means that rsync will only copy the contents of your DOCK_Tutorial folders. If you want to copy the DOCK_Tutorial folder itself, as well as its contents, then remove the trailing /.
To copy newer files from Seawulf back to Herbie, do the following from Seawulf:
rsync -arv /nfs/user03/username/DOCK_Tutorial/ username@compute.mathlab.sunysb.edu:/home/username/DOCK_Tutorial
You can apply this same strategy to then sync files with your home computer as well. Use the following format:
rsync -arv source target
- Note: this is easiest if you are using a Linux or Mac computer at home.
Deleting jobs
If you make a mistake and need to delete a job from the queue, first list all your queued or running jobs:
qstat -u username
- IMPORTANT: If you get no output from qstat, it means that whatever jobs you have submitted are done!
If you do have jobs running or waiting in the queue, qstat will output a list that includes their job id(s). Find the job id for the one you want to delete, and do:
qdel jobid