Difference between revisions of "2014 DOCK tutorial with HIV Protease"
Stonybrook (talk | contribs) (→IV. Generating Box and Grid) |
Stonybrook (talk | contribs) (→VI. Virtual Screening) |
||
| (55 intermediate revisions by 2 users not shown) | |||
| Line 42: | Line 42: | ||
Put the 1HVR PDB file in 00.file/folder. If you are in the 00.files/directory, then tap the command: | Put the 1HVR PDB file in 00.file/folder. If you are in the 00.files/directory, then tap the command: | ||
cp ~/Downloads/1HVR.pdb ./ | cp ~/Downloads/1HVR.pdb ./ | ||
| − | When you are preparing | + | When you are preparing your PDB files, you have to make some modifications on your original file. Make a copy of your original PDB file, and rename it as "1HVR.modified.pdb" in the 00.files/. Here we changed the atom name from "CSO" to "CYS" and deleted two lines "OD" and "HD". Here is an example to change all "CSO" to "CYS": |
| + | :%s/CSO/CYS/gc | ||
And then we will create 4 files in 01.dockprep/ directory: | And then we will create 4 files in 01.dockprep/ directory: | ||
1HVR.dockprep.mol2 | 1HVR.dockprep.mol2 | ||
| Line 50: | Line 51: | ||
''Create the dockprep file'' | ''Create the dockprep file'' | ||
| − | For the "1HVR.dockprep.mol2" file: open the 1HVR.modified.pdb in Chimera; delete the water molecules; delete the original hydrogen atoms; add the charge | + | For the "1HVR.dockprep.mol2" file: open the 1HVR.modified.pdb in Chimera; delete the water molecules ; delete the original hydrogen atoms; add the charge by go to Tools->structure editing->add charge ''(Note when adding the charge to the ligand, you can choose AMBER ff99SB as the charge model and chose gasteiger as the charge method. In this 1HVR case, we set Net Charge to 0. You may have to consider the chemistry of the ligand when assigning a charge state)''. Add the hydrogen atoms manually by Tools->structure editing->Add H . Or you can do all of the above by clicking Tools -> Structure Editing -> Dock Prep. |
| − | Note when adding the charge to the ligand, you can choose AMBER ff99SB as the charge model and chose gasteiger as the charge method. In this 1HVR case, we set Net Charge to 0. | + | |
Finally save the file as a mol2 format in Chimera. | Finally save the file as a mol2 format in Chimera. | ||
''Create the ligand file'' | ''Create the ligand file'' | ||
| − | To create the ligand file: Open 1HVR.dockprep.mol2 in Chimera and select the " | + | To create the ligand file: Open 1HVR.dockprep.mol2 in Chimera and select the ligand chain "XK2", then Select->invert, then go to Action->Atoms/bonds->Delete |
''Create the receptor file'' | ''Create the receptor file'' | ||
| − | Select the residue | + | Select the ligand residue "XK2" and delete it and save it as a mol2 file. |
''Create the receptor without hydrogen atoms'' | ''Create the receptor without hydrogen atoms'' | ||
| − | Open the "1HVR.receptor.mol2" file and delete all of the hydrogen atoms in Chimera and | + | Open the "1HVR.receptor.mol2" file and delete all of the hydrogen atoms in Chimera by go to Select->Chemistry->Element->H and then Action->Atoms/bonds->Delete.Save it as a pdb file. |
==III. Generating Receptor Surface and Spheres== | ==III. Generating Receptor Surface and Spheres== | ||
| Line 127: | Line 128: | ||
'''3.''' ''(optional)'' To look at the spheres generated, you need to put them into PDB format. | '''3.''' ''(optional)'' To look at the spheres generated, you need to put them into PDB format. | ||
| − | Run '''showsphere''', by typing the | + | Run '''showsphere''', by typing the following into the terminal: |
showsphere | showsphere | ||
| Line 181: | Line 182: | ||
==IV. Generating Box and Grid== | ==IV. Generating Box and Grid== | ||
| − | + | === Box Generation=== | |
Mosavverul Arkin | Mosavverul Arkin | ||
| − | * Make a new directory and name it: 03.box-grid/ | + | * Make a new directory and name it: ''03.box-grid/'' |
mkdir 03.box-grid | mkdir 03.box-grid | ||
| − | * Make a new file in this directory and name it showbox.in | + | * Make a new file in this directory and name it ''showbox.in'' |
vim showbox.in | vim showbox.in | ||
| − | * This will automatically open the file showbox.in. Edit the file showbox.in as follows: | + | * This will automatically open the file ''showbox.in''. Edit the file ''showbox.in'' as follows: |
| − | Y #Yes, generate a box | + | Y # Yes, generate a box |
| − | 8.0 #Size of the box in Angstroms | + | 8.0 # Size of the box in Angstroms |
| − | ../02. | + | ../02.surface-spheres/selected_spheres.sph # Sphere.sph file |
| − | 1 #Cluster number | + | 1 # Cluster number |
| − | 1HVR.box.pdb #Name of the output file | + | 1HVR.box.pdb # Name of the output file |
| − | * Save the file | + | * Save the file using the command: |
:wq | :wq | ||
| − | + | in ''esc'' mode | |
* Run the command: | * Run the command: | ||
showbox < showbox.in | showbox < showbox.in | ||
| Line 208: | Line 209: | ||
[[Image:1HVR_receptor_surface_2014.png|thumb|center|375px|Receptor surface with generated box]] | [[Image:1HVR_receptor_surface_2014.png|thumb|center|375px|Receptor surface with generated box]] | ||
| − | + | === Grid computing=== | |
| − | + | We now create the energy scoring grid file '' *.nrg '' using the program named ''grid''. | |
| − | + | ||
| − | + | First you need to make sure that DOCK6 is installed, you can determine the path of DOCK installation by this command | |
| − | + | which dock6 | |
| − | + | from this location, make sure you have these three files and copy them to your local ~00.files/folder | |
| − | + | ../parameters/vdw_AMBER_parm99.defn | |
| − | + | ../parameters/flex.defn | |
| − | + | ../parameters/flex_drive.tbl | |
| − | + | * Make a new file in the same directory (''03.box-grid/'') and name it: ''grid.in'' | |
| − | + | touch grid.in | |
| − | + | grid -i grid.in | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | This will automatically open the file grid.in. Edit the file ''grid.in'' with the parameters and values listed in the table below. | ||
| + | The program ''grid'' also computes a bump grid ''*.bmp'' file which identifies whether a ligand atom is in severe steric overlap with a receptor atom. For more information please refer to [http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm#Grid DOCK 6.6 Users Manual] | ||
| + | {| border="1" cellpadding="5" cellspacing="0" align="center" | ||
| + | |+ '''grid.in''' | ||
| + | |- | ||
| + | ! style="background: #efefef;" | Parameter | ||
| + | ! style="background: #efefef;" | Value | ||
| + | ! style="background: #efefef;" | Description | ||
| + | |- | ||
| + | | compute_grids | ||
| + | | yes | ||
| + | | compute scoring grids (yes) | ||
| + | |- | ||
| + | | grid_spacing | ||
| + | | 0.4 | ||
| + | | distance between grid points along each axis (in Å). | ||
| + | |- | ||
| + | | output_molecule | ||
| + | | no | ||
| + | | write up coordinates of the receptor into a new file | ||
| + | |- | ||
| + | | contact_score | ||
| + | | no | ||
| + | | compute contact grid? default is no | ||
| + | |- | ||
| + | | energy_score | ||
| + | | yes | ||
| + | | compute energy score? yes - we are using this method to compute force fields on probes | ||
| + | |- | ||
| + | | energy_cutoff_distance | ||
| + | | 9999 | ||
| + | | the max distance between atoms for the energy contribution to be computed | ||
| + | |- | ||
| + | | atom_model | ||
| + | | a | ||
| + | | atom_model u means united atom model where atoms are attached to hydrogens, and a stands for all-atom model, where hydrogens on carbons are treated separately | ||
| + | |- | ||
| + | | attractive_exponent | ||
| + | | 6 | ||
| + | | attractive component stands for exponent of the attractive LJ term in VDW potential | ||
| + | |- | ||
| + | | repulsive_exponent | ||
| + | | 9 | ||
| + | | repulsive component stands for exponent in the repulsive LJ term in VDW potential | ||
| + | |- | ||
| + | | distance_dielectric | ||
| + | | yes | ||
| + | | distance dielectric stands for the dielectric constant to be linearly dependent on distance | ||
| + | |- | ||
| + | | dielectric_factor | ||
| + | | 4 | ||
| + | | distance dielectric factor is the coefficient of the dielectric | ||
| + | |- | ||
| + | | bump_filter | ||
| + | | yes | ||
| + | | bump filter flag determines if we want to screen orientation for clashes before scoring and minimization | ||
| + | |- | ||
| + | | bump_overlap | ||
| + | | 0.75 | ||
| + | | bump_overlap stands for the fraction of allowed overlap where 1 corresponds to no allowed overlap and 0 corresponds to full overlap being permitted. | ||
| + | |- | ||
| + | | receptor_file | ||
| + | | ../01.dockprep/1HVR.receptor.mol2 | ||
| + | | our receptor file | ||
| + | |- | ||
| + | | box_file | ||
| + | | 1HVR.box.pdb | ||
| + | | the box file we generated in the Box section | ||
| + | |- | ||
| + | | vdw_definition_file | ||
| + | | ../00.files/vdw_AMBER_parm99.defn | ||
| + | | van der Waals parameters file | ||
| + | |- | ||
| + | | style="border-bottom: 3px solid grey;" | score_grid_prefix | ||
| + | | style="border-bottom: 3px solid grey;" | 1HVR.grid | ||
| + | | style="border-bottom: 3px solid grey;" | prefix for the grid file name; all the extensions will be generated automatically. | ||
| + | |- | ||
| − | + | |} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==V. Docking a Single Molecule for Pose Reproduction== | ==V. Docking a Single Molecule for Pose Reproduction== | ||
| Line 260: | Line 308: | ||
Rigid Ligand Docking is usually used to check the validity of docking program and user-defined parameters because it's much faster than flexible docking. It also helps users to identify any mistakes made during the preparation of receptor and ligand. In our case, we know the experimental structure of a receptor-ligand complex. So if we dock the ligand back to the ligand-free receptor. The pose predicted by DOCK should be very close to the experimental one. Otherwise, users should examine the parameters and all the files involved. | Rigid Ligand Docking is usually used to check the validity of docking program and user-defined parameters because it's much faster than flexible docking. It also helps users to identify any mistakes made during the preparation of receptor and ligand. In our case, we know the experimental structure of a receptor-ligand complex. So if we dock the ligand back to the ligand-free receptor. The pose predicted by DOCK should be very close to the experimental one. Otherwise, users should examine the parameters and all the files involved. | ||
| + | The following should be performed in 04.dock/file | ||
| + | |||
| + | First create an empty file and execute DOCK6. The dock.in is the name of the input file(your input file may have another name). | ||
| + | touch dock.in | ||
| + | dock6 -i dock.in | ||
Here is the input for DOCK6 | Here is the input for DOCK6 | ||
ligand_atom_file ../01.dockprep/1HVR.ligand.mol2 | ligand_atom_file ../01.dockprep/1HVR.ligand.mol2 | ||
| Line 309: | Line 362: | ||
simplex_coefficient_restraint 10.0 | simplex_coefficient_restraint 10.0 | ||
atom_model all | atom_model all | ||
| − | vdw_defn_file ../ | + | vdw_defn_file ../00.files/vdw_AMBER_parm99.defn |
flex_defn_file ../00.files/flex.defn | flex_defn_file ../00.files/flex.defn | ||
flex_drive_file ../00.files/flex_drive.tbl | flex_drive_file ../00.files/flex_drive.tbl | ||
| Line 320: | Line 373: | ||
Note that "flexible_ligand" is turned off. "minimize_ligand" has to be on. | Note that "flexible_ligand" is turned off. "minimize_ligand" has to be on. | ||
| + | |||
| + | |||
| + | You will have a file named "1HVR.out_scored.mol2" file in the directory where you run dock6. This file have the coordinates of ligand as "1HVR.ligand.mol2". Moreover, it also has energy score and rmsd value. You can open the pdb file of the complex in CHIMERA and open viewdock in Tool->surface->binding analysis. In viewdock, open the "1HVR.out_scored.mol2" file and viewdock will superimpose the predicted ligand pose onto the complex. You can have a visualized examination of the predicted pose. | ||
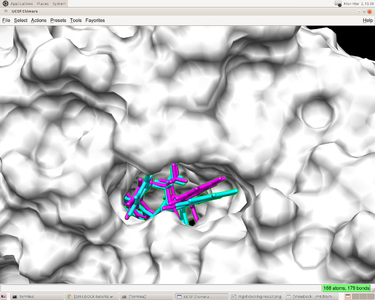
[[Image:Rigid docking result.png|thumb|center|375px|Rigid docking result;RMSD = 0.9848; green: experiment; purple: predicted]] | [[Image:Rigid docking result.png|thumb|center|375px|Rigid docking result;RMSD = 0.9848; green: experiment; purple: predicted]] | ||
| − | The predicted pose and the experimental pose have an RMSD less than 2.000 which means the prediction reproduced the result from experiment. | + | The predicted pose and the experimental pose have an RMSD less than 2.000 which means the prediction reproduced the result from experiment. The preparation of receptor and ligand is correct and we can go to the next steps(such as flexible ligand docking and virtual screening) with some minor modification of this input file. |
'''Flexible Ligand DOCKing''' | '''Flexible Ligand DOCKing''' | ||
| + | |||
| + | For flexible docking, some parameters need to be changed, and these changes will generate several additional questions to the user. Some changes of parameters are shown below: | ||
| + | calculate_rmsd yes | ||
| + | flexible_ligand yes | ||
| + | use_internal_energy yes | ||
| + | minimize_ligand yes | ||
| + | num_scored_conformers 10 | ||
| + | After the changes are made, execute the DOCK6: | ||
| + | dock6 -i dock.in | ||
| + | Several additional questions will be asked, and answer them according to the table below: | ||
| + | |||
| + | ligand_atom_file ../01.dockprep/1HVR.ligand.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../01.dockprep/1HVR.ligand.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../02.surface-spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | flexible_ligand yes | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 100 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | contact_score_secondary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_secondary no | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../03.box-grod/1HVR.grid | ||
| + | multigrid_score_secondary no | ||
| + | dock3.5_score_secondary no | ||
| + | continuous_score_secondary no | ||
| + | descriptor_score_secondary no | ||
| + | gbsa_zou_score_secondary no | ||
| + | gbsa_hawkins_score_secondary no | ||
| + | SASA_descriptor_score_secondary no | ||
| + | amber_score_secondary no | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 500 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min yes | ||
| + | simplex_coefficient_restraint 10.0 | ||
| + | atom_model all | ||
| + | vdw_defn_file ../00.files/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file ../00.files/flex.defn | ||
| + | flex_drive_file ../00.files/flex_drive.tbl | ||
| + | ligand_outfile_prefix 1HVR.output | ||
| + | write_orientations no | ||
| + | num_scored_conformers 100 | ||
| + | write_conformations no | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | Note that "flexible_ligand" is on. | ||
In the case of flexible ligand DOCKing, the ligand is allowed to be flexible. This type of docking allows the ligand to structurally rearrange in response to the receptor. Initially the largest rigid substructure of the ligand is identified; then the flexible layers are identified. The orientations are ranked according to their score, and are grouped by root mean squared deviation (RMSD). To run the docking calculation, use the program dock6 that is distributed with DOCK in the /bin directory. You need to generate an input file by answering questions interactively or manually via text file. The program will generate an output file summarizing the parameters used in the run, any warning or error messages, and summary information about the best scoring pose. It will also produce a structure (.mol2) file, containing the geometric coordinates of the best pose as well as a summary of the interaction energy of that pose. | In the case of flexible ligand DOCKing, the ligand is allowed to be flexible. This type of docking allows the ligand to structurally rearrange in response to the receptor. Initially the largest rigid substructure of the ligand is identified; then the flexible layers are identified. The orientations are ranked according to their score, and are grouped by root mean squared deviation (RMSD). To run the docking calculation, use the program dock6 that is distributed with DOCK in the /bin directory. You need to generate an input file by answering questions interactively or manually via text file. The program will generate an output file summarizing the parameters used in the run, any warning or error messages, and summary information about the best scoring pose. It will also produce a structure (.mol2) file, containing the geometric coordinates of the best pose as well as a summary of the interaction energy of that pose. | ||
| Line 351: | Line 494: | ||
mv dock.in mini-virtual-screen.in | mv dock.in mini-virtual-screen.in | ||
| − | + | The content of mini-virtual-screen.in is as follows: | |
ligand_atom_file HIVPR.ligands.005.mol2 | ligand_atom_file HIVPR.ligands.005.mol2 | ||
| Line 422: | Line 565: | ||
'''Analyzing the Results''' | '''Analyzing the Results''' | ||
| − | After | + | After performinging virtual screening on local machine, a file named 1HVR.output_scored.mol2 is obtained, which contains successfully docked ligands, including grid score, internal energy, etc. |
| Line 428: | Line 571: | ||
########## Grid Score: -25.904999 | ########## Grid Score: -25.904999 | ||
########## Grid_vdw: -25.904999 | ########## Grid_vdw: -25.904999 | ||
| − | ########## | + | ########## Grid_es: 0.000000 |
########## Int_energy: 0.062399 | ########## Int_energy: 0.062399 | ||
| − | |||
@<TRIPOS>MOLECULE | @<TRIPOS>MOLECULE | ||
NONAME | NONAME | ||
| Line 436: | Line 578: | ||
SMALL | SMALL | ||
NO_CHARGES | NO_CHARGES | ||
| − | |||
@<TRIPOS>ATOM | @<TRIPOS>ATOM | ||
1 O -6.5962 19.4021 23.0405 O.2 1 <1> 0.0000 | 1 O -6.5962 19.4021 23.0405 O.2 1 <1> 0.0000 | ||
| Line 480: | Line 621: | ||
20 19 20 1 | 20 19 20 1 | ||
| − | + | When virtual screening with a larger ligand database runs completely in Seawulf, files 1HVR.output_scored.mol2 which contains the mol2 files of all successfully docked ligands, virtual_screen.out which contains the dock results of successfully docked ligands, and virtual_screen.out.1 to virtual_screen.out.3 which contain dock results from different nodes you use (the number is related with the number of nodes you use, here we use 4 nodes) are produced. One of the successfully docked ligands is as follows: | |
Molecule: CHEMBL171551 | Molecule: CHEMBL171551 | ||
| − | + | Anchors: 3 | |
| − | |||
Orientations: 3000 | Orientations: 3000 | ||
Conformations: 359 | Conformations: 359 | ||
| − | + | Grid Score: -60.092117 | |
| − | |||
Grid_vdw: -62.332535 | Grid_vdw: -62.332535 | ||
| − | + | Grid_es: 2.240418 | |
Int_energy: 13.718960 | Int_energy: 13.718960 | ||
| − | These files should copied to local computers if we want to use Chimera to visually check the results. In Seawulf, move to 06.virtual-screen/ directory, | + | These files should be copied to local computers if we want to use Chimera to visually check the results. In Seawulf, move to 06.virtual-screen/ directory, |
| − | + | scp –r username@herbie.mathlab.sunysb.edu:~/AMS536/dock_tutorial/ | |
| − | In herbie, open the receptor 1HVR.receptor.mol2 and reference ligand 1HVR.ligand.mol2 | + | In herbie, open the receptor 1HVR.receptor.mol2 and reference ligand 1HVR.ligand.mol2 and then how the ligands fitting the receptor can be checked by ViewDock function in Chimera. Go to Tools -> Surface/Binding Analysis -> ViewDock and choose 1HVR.output_scored.mol2.In the pup-up window, we can go to Column ->show -> Grid Score to show the grid score. |
==VII. Running DOCK in Parallel on Seawulf== | ==VII. Running DOCK in Parallel on Seawulf== | ||
| Line 557: | Line 696: | ||
qstat #the whole list of submitted jobs in queue | qstat #the whole list of submitted jobs in queue | ||
| − | + | qstat -u username #the whole list of jobs in queue you submitted | |
| Line 564: | Line 703: | ||
==VIII. Frequently Encountered Problems== | ==VIII. Frequently Encountered Problems== | ||
| − | You should always be clear about what the input file and output file of one program are and what format they are. | + | * You should always be clear about what the input file and output file of one program are and what format they are. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * Sometimes, it's easy to make mistakes if you are confused about your current location, or "path", so make sure you know that all the time. | |
| − | + | * The most common problem experienced in UNIX is having the wrong file path. '''ALWAYS CHECK TO MAKE SURE THAT YOUR FILE PATH IS CORRECT''' This can be facilitated with the Tab button which will auto complete what you have started to type. | |
| − | + | * Make sure you are opening the correct files for each visualization step (e.g. opening the receptor file with the ligand still there may make analyzing your docking results a bit more challenging). | |
| − | + | * Use the auto-fill (tab) function but double check to make sure the correct file was auto-filled (e.g. if you type ''1H'' then hit tab you might get ''1HVR_selected_spheres'' but the file you want actually is ''1HVR_selected_spheres.sph''). | |
| − | + | * For users with no UNIX experience and limited programming experience - it's a good idea to run through a couple tutorials before you start using UNIX. Even if you type in the commands without fully understanding their implications, become familiar with the basic commands and shortcuts. It will help a lot when you actually start using UNIX! | |
| − | + | * If you are not so familiar with UNIX environment, it is better for you to practice the tutorial with basic commands: http://www.ee.surrey.ac.uk/Teaching/Unix/ | |
| − | + | * When you are trying a command, but have a typo (so this command will not work), we can always go back to the last command by press UP arrow button go back to the command that just typed and modify that command. | |
| − | + | * Insert button can allows us to retype the command instead of rewrite it. | |
| − | + | * Example for writing text, writing a command, and importing an image: | |
Write some text here.. | Write some text here.. | ||
Latest revision as of 18:05, 17 December 2014
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2013, using DOCK v6.6.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
HIV Protease
HIV protease (HIVPR) is a protein involved in viral maturation during the life cycle of HIV. HIVPR is an approximately 22 kDa homodimer with 99 residues per chain. Inhibition of this protein has been shown to be an effective form of treatment of HIV. Currently-available HIVPR inhibitors generally take the form of a symmetric cyclic urea compound. For more information, see Lam et al.
Organizing Directories
While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify. For this tutorial, we will use something similar to the following:
~username/AMS536/dock-tutorial/00.files/
/01.dockprep/
/02.surface-spheres/
/03.box-grid/
/04.dock/
/05.mini-virtual-screen/
/06.virtual-screen/
In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is the same as the PDB code, '1HVR'. The following sections in this tutorial will adhere to this directory structure / naming scheme.
II. Preparing the Receptor and Ligand
Downloading the PDB Structure (1HVR)
Go to PDB (Protein Data Bank) website (http://www.rcsb.org/pdb/home/home.do) enter the protein ID (1HVR), search for the PDB file and download it as a text form.
Preparing the ligand and receptor in Chimera
Put the 1HVR PDB file in 00.file/folder. If you are in the 00.files/directory, then tap the command:
cp ~/Downloads/1HVR.pdb ./
When you are preparing your PDB files, you have to make some modifications on your original file. Make a copy of your original PDB file, and rename it as "1HVR.modified.pdb" in the 00.files/. Here we changed the atom name from "CSO" to "CYS" and deleted two lines "OD" and "HD". Here is an example to change all "CSO" to "CYS":
:%s/CSO/CYS/gc
And then we will create 4 files in 01.dockprep/ directory:
1HVR.dockprep.mol2 1HVR.ligand.mol2 1HVR.receptor.mol2 1HVR.receptor.noH.pdb
Create the dockprep file
For the "1HVR.dockprep.mol2" file: open the 1HVR.modified.pdb in Chimera; delete the water molecules ; delete the original hydrogen atoms; add the charge by go to Tools->structure editing->add charge (Note when adding the charge to the ligand, you can choose AMBER ff99SB as the charge model and chose gasteiger as the charge method. In this 1HVR case, we set Net Charge to 0. You may have to consider the chemistry of the ligand when assigning a charge state). Add the hydrogen atoms manually by Tools->structure editing->Add H . Or you can do all of the above by clicking Tools -> Structure Editing -> Dock Prep.
Finally save the file as a mol2 format in Chimera.
Create the ligand file
To create the ligand file: Open 1HVR.dockprep.mol2 in Chimera and select the ligand chain "XK2", then Select->invert, then go to Action->Atoms/bonds->Delete
Create the receptor file
Select the ligand residue "XK2" and delete it and save it as a mol2 file.
Create the receptor without hydrogen atoms
Open the "1HVR.receptor.mol2" file and delete all of the hydrogen atoms in Chimera by go to Select->Chemistry->Element->H and then Action->Atoms/bonds->Delete.Save it as a pdb file.
III. Generating Receptor Surface and Spheres
Generating the Receptor Surface
Check to make sure 02.surface-spheres directory exists under dock-tutorial. If not then make the following directory:
mkdir 02.surface-sphgen cd 02.surface-sphgen
The following steps will be carried out to generate the receptor surface using Chimera:
Open Chimera by simply typing chimera into the terminal window
| Go File -> Open and choose the PDB file of the protein containing no hydrogens (1HVR.receptor.noH.pdb) from 01.dockprep
| Further, Actions -> Surface -> Show
| Go Tools -> Structure Editing -> Write DMS in order to obtain a dms file, which we will need to place spheres
| In the new window save the surface as 1HVR.receptor.dms
Placing Spheres
We will be using SPHGEN to generate spheres: see the DOCK online owners manual for additional information:
<http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm>
The following steps will be used to place the spheres on the receptor surface:
1. Create a file called INSPH and fill it out as follows, then save it. This input file tells SPHGEN what to do, details of each line are below:
1HVR.receptor.dms R X 0.0 4.0 1.4 1HVR.receptor.sph
Input File Details:
1HVR.receptor.dms - surface file from the previous step R - tells SPHGEN to place spheres either outside of the surface (R) or inside the surface (L) X - tells SPHGEN the subset of surface points to be used (X=all points) 0.0 - prevents generation of large spheres with close surface contacts(defalut=0.0) 4.0 - maximum sphere radius in angstroms (default=4.0) 1.4 - minimum sphere radius in angstroms (default=radius of probe) 1HVR.receptor.sph - clustered spheres file that we want to generate
2. Run the sphgen program from the terminal:
sphgen -i INSPH -o OUTSPH
-i tells sphgen where the input file INSPH is INSPH tells sphgen what to do -o tells sphgen what to call the oputput file OUTSPH is the output file containing the sphere information
3. (optional) To look at the spheres generated, you need to put them into PDB format.
Run showsphere, by typing the following into the terminal:
showsphere
You will be prompted with the following questions:
Enter name of sphere cluster file:
1HVR.receptor.sph
Enter cluster number to process (<0 = all):
-1
Generate surfaces as well as pdb files (<N>/Y)?
N
Enter name for output file prefix:
output_spheres
Process cluster 0 (contains ALL spheres) (<N>/Y)?
N
You can then open the receptor file in Chimera as well as the output_spheres.pdb file and should see many spheres placed all over the receptor surface.
4. Run the sphere_selector program from in the terminal to select spheres of interest (note: not all of the spheres in the previous image are in the active site so we want to eliminate them):
sphere_selector 1HVR.receptor.sph ../01.dockprep/1HVR.ligand.mol2 8.0
This program goes to select the spheres within a user-defined radius (8.0 here) of a target molecule from a previously obtained file: 1HVR.receptor.sph. In turn, a new file selected_spheres.sph will be generated.
5. Run showsphere to visualize the spheres:
showsphere
When prompted on the command line, answering the questions as follows:
Enter name of sphere cluster file:
selected_spheres.sph
Enter cluster number to process (<0 = all):
-1
Generate surfaces as well as pdb files (<N>/Y)?
N
Enter name for output file prefix:
output_spheres_selected
Process cluster 0 (contains ALL spheres) (<N>/Y)?
N
- Launch Chimera, choose File -> Open, choose 1HVR.receptor.noH.pdb
- Go File -> Open, choosing output_spheres_selected.pdb
- Go Select -> Residue -> SPH
- Finally, Actions -> Atoms/Bonds -> sphere
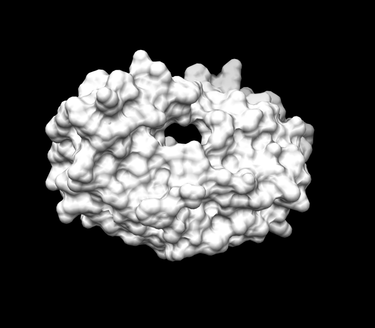
The final image should look similar to the example below:
IV. Generating Box and Grid
Box Generation
Mosavverul Arkin
- Make a new directory and name it: 03.box-grid/
mkdir 03.box-grid
- Make a new file in this directory and name it showbox.in
vim showbox.in
- This will automatically open the file showbox.in. Edit the file showbox.in as follows:
Y # Yes, generate a box 8.0 # Size of the box in Angstroms ../02.surface-spheres/selected_spheres.sph # Sphere.sph file 1 # Cluster number 1HVR.box.pdb # Name of the output file
- Save the file using the command:
:wq
in esc mode
- Run the command:
showbox < showbox.in
Grid computing
We now create the energy scoring grid file *.nrg using the program named grid.
First you need to make sure that DOCK6 is installed, you can determine the path of DOCK installation by this command
which dock6
from this location, make sure you have these three files and copy them to your local ~00.files/folder
../parameters/vdw_AMBER_parm99.defn ../parameters/flex.defn ../parameters/flex_drive.tbl
- Make a new file in the same directory (03.box-grid/) and name it: grid.in
touch grid.in grid -i grid.in
This will automatically open the file grid.in. Edit the file grid.in with the parameters and values listed in the table below. The program grid also computes a bump grid *.bmp file which identifies whether a ligand atom is in severe steric overlap with a receptor atom. For more information please refer to DOCK 6.6 Users Manual
| Parameter | Value | Description |
|---|---|---|
| compute_grids | yes | compute scoring grids (yes) |
| grid_spacing | 0.4 | distance between grid points along each axis (in Å). |
| output_molecule | no | write up coordinates of the receptor into a new file |
| contact_score | no | compute contact grid? default is no |
| energy_score | yes | compute energy score? yes - we are using this method to compute force fields on probes |
| energy_cutoff_distance | 9999 | the max distance between atoms for the energy contribution to be computed |
| atom_model | a | atom_model u means united atom model where atoms are attached to hydrogens, and a stands for all-atom model, where hydrogens on carbons are treated separately |
| attractive_exponent | 6 | attractive component stands for exponent of the attractive LJ term in VDW potential |
| repulsive_exponent | 9 | repulsive component stands for exponent in the repulsive LJ term in VDW potential |
| distance_dielectric | yes | distance dielectric stands for the dielectric constant to be linearly dependent on distance |
| dielectric_factor | 4 | distance dielectric factor is the coefficient of the dielectric |
| bump_filter | yes | bump filter flag determines if we want to screen orientation for clashes before scoring and minimization |
| bump_overlap | 0.75 | bump_overlap stands for the fraction of allowed overlap where 1 corresponds to no allowed overlap and 0 corresponds to full overlap being permitted. |
| receptor_file | ../01.dockprep/1HVR.receptor.mol2 | our receptor file |
| box_file | 1HVR.box.pdb | the box file we generated in the Box section |
| vdw_definition_file | ../00.files/vdw_AMBER_parm99.defn | van der Waals parameters file |
| score_grid_prefix | 1HVR.grid | prefix for the grid file name; all the extensions will be generated automatically. |
V. Docking a Single Molecule for Pose Reproduction
Rigid Ligand DOCKing
Rigid Ligand Docking is usually used to check the validity of docking program and user-defined parameters because it's much faster than flexible docking. It also helps users to identify any mistakes made during the preparation of receptor and ligand. In our case, we know the experimental structure of a receptor-ligand complex. So if we dock the ligand back to the ligand-free receptor. The pose predicted by DOCK should be very close to the experimental one. Otherwise, users should examine the parameters and all the files involved.
The following should be performed in 04.dock/file
First create an empty file and execute DOCK6. The dock.in is the name of the input file(your input file may have another name).
touch dock.in dock6 -i dock.in
Here is the input for DOCK6
ligand_atom_file ../01.dockprep/1HVR.ligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1HVR.ligand.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no use_internal_energy no flexible_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1HVR.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix 1HVR.out write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations no rank_ligands no
Note that "flexible_ligand" is turned off. "minimize_ligand" has to be on.
You will have a file named "1HVR.out_scored.mol2" file in the directory where you run dock6. This file have the coordinates of ligand as "1HVR.ligand.mol2". Moreover, it also has energy score and rmsd value. You can open the pdb file of the complex in CHIMERA and open viewdock in Tool->surface->binding analysis. In viewdock, open the "1HVR.out_scored.mol2" file and viewdock will superimpose the predicted ligand pose onto the complex. You can have a visualized examination of the predicted pose.
The predicted pose and the experimental pose have an RMSD less than 2.000 which means the prediction reproduced the result from experiment. The preparation of receptor and ligand is correct and we can go to the next steps(such as flexible ligand docking and virtual screening) with some minor modification of this input file.
Flexible Ligand DOCKing
For flexible docking, some parameters need to be changed, and these changes will generate several additional questions to the user. Some changes of parameters are shown below:
calculate_rmsd yes flexible_ligand yes use_internal_energy yes minimize_ligand yes num_scored_conformers 10
After the changes are made, execute the DOCK6:
dock6 -i dock.in
Several additional questions will be asked, and answer them according to the table below:
ligand_atom_file ../01.dockprep/1HVR.ligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1HVR.ligand.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no use_internal_energy yes internal_energy_rep_exp 12 flexible_ligand yes user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 100 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 use_clash_overlap no write_growth_tree no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grod/1HVR.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix 1HVR.output write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Note that "flexible_ligand" is on.
In the case of flexible ligand DOCKing, the ligand is allowed to be flexible. This type of docking allows the ligand to structurally rearrange in response to the receptor. Initially the largest rigid substructure of the ligand is identified; then the flexible layers are identified. The orientations are ranked according to their score, and are grouped by root mean squared deviation (RMSD). To run the docking calculation, use the program dock6 that is distributed with DOCK in the /bin directory. You need to generate an input file by answering questions interactively or manually via text file. The program will generate an output file summarizing the parameters used in the run, any warning or error messages, and summary information about the best scoring pose. It will also produce a structure (.mol2) file, containing the geometric coordinates of the best pose as well as a summary of the interaction energy of that pose.
VI. Virtual Screening
Virtual Screening Introduction
Virtual screening is a method used to predict most favorable ligand binding to a target protein within a ligand database. It also allows for comparison of both qualitative (e.g. position in binding site) and quantitative (e.g. grid scores, internal energy) data pertaining to the each screened ligand with the originally docked molecule.
To perform virtual screening, HIVPR.ligands.005.mol2, a mol2 file that contains 5 small molecules will be the virtual library based on the receptor 1HVR. This is a reasonable computational cost for a quick search, so we can conduct it on our own computer. After that, virtual screening can be conducted within a larger database, HIVPR.ligands.100.mol2, which contains 100 ligands. Since the computational cost of virtual screening is much higher, it is better for us to run it on Seawulf.
Running Virtual Screening
Before running virtual screening, a new directory 05.mini-virtual-screen is created and ligand files HIVPR.ligands.005.mol2 and HIVPR.ligands.100.mol2 are copied from Wjallen. Since most parameters of dock.in and new virtual screening input are the same, we can make few modifications to dock.in script but rename it as mini-virtual-screen.in.
mkdir 05.mini-virtual-screen cd 05.mini-virtual-screen cp ~wjallen/AMS536/multi-mol2/files/HIVPR.ligands.005.mol2 ./ cp ../04.dock/dock.in/ ./ mv dock.in mini-virtual-screen.in
The content of mini-virtual-screen.in is as follows:
ligand_atom_file HIVPR.ligands.005.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no use_internal_energy yes internal_energy_rep_exp 12 flexible_ligand yes user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100 use_clash_overlap no write_growth_tree no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1HVR.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 1000 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file ../00.files/vdw_AMBER_parm99.defn flex_defn_file ../00.files/flex.defn flex_drive_file ../00.files/flex_drive.tbl ligand_outfile_prefix 1HVR.output write_orientations no num_scored_conformers 100 rank_ligands no
Analyzing the Results
After performinging virtual screening on local machine, a file named 1HVR.output_scored.mol2 is obtained, which contains successfully docked ligands, including grid score, internal energy, etc.
########## Name: NONAME
########## Grid Score: -25.904999
########## Grid_vdw: -25.904999
########## Grid_es: 0.000000
########## Int_energy: 0.062399
@<TRIPOS>MOLECULE
NONAME
20 20 1 0 0
SMALL
NO_CHARGES
@<TRIPOS>ATOM
1 O -6.5962 19.4021 23.0405 O.2 1 <1> 0.0000
2 C -6.8355 18.4953 22.2343 C.2 1 <1> 0.0000
3 N -7.8272 17.5802 22.4935 N.am 1 <1> 0.0000
4 H -8.3181 17.7085 23.3731 H 1 <1> 0.0000
5 C -8.2659 16.4500 21.6933 C.3 1 <1> 0.0000
6 H -9.1047 15.9926 22.2333 H 1 <1> 0.0000
7 H -7.4773 15.6904 21.6784 H 1 <1> 0.0000
8 C -8.7214 16.8444 20.2915 C.3 1 <1> 0.0000
9 H -9.4627 17.6481 20.3875 H 1 <1> 0.0000
10 O -9.4130 15.7379 19.7151 O.3 1 <1> 0.0000
11 H -10.1762 15.5384 20.2849 H 1 <1> 0.0000
12 C -7.5968 17.3421 19.3635 C.3 1 <1> 0.0000
13 H -7.8639 18.3502 19.0239 H 1 <1> 0.0000
14 O -7.5032 16.5445 18.1836 O.3 1 <1> 0.0000
15 H -8.3854 16.5076 17.7791 H 1 <1> 0.0000
16 C -6.2182 17.4085 20.0113 C.3 1 <1> 0.0000
17 H -5.4732 17.6806 19.2547 H 1 <1> 0.0000
18 H -5.8956 16.4394 20.4044 H 1 <1> 0.0000
19 N -6.1245 18.4045 21.0622 N.am 1 <1> 0.0000
20 H -5.4272 19.1287 20.9184 H 1 <1> 0.0000
@<TRIPOS>BOND
1 1 2 2
2 2 3 am
3 2 19 am
4 3 4 1
5 3 5 1
6 5 6 1
7 5 7 1
8 5 8 1
9 8 9 1
10 8 10 1
11 8 12 1
12 10 11 1
13 12 13 1
14 12 14 1
15 12 16 1
16 14 15 1
17 16 17 1
18 16 18 1
19 16 19 1
20 19 20 1
When virtual screening with a larger ligand database runs completely in Seawulf, files 1HVR.output_scored.mol2 which contains the mol2 files of all successfully docked ligands, virtual_screen.out which contains the dock results of successfully docked ligands, and virtual_screen.out.1 to virtual_screen.out.3 which contain dock results from different nodes you use (the number is related with the number of nodes you use, here we use 4 nodes) are produced. One of the successfully docked ligands is as follows:
Molecule: CHEMBL171551
Anchors: 3
Orientations: 3000
Conformations: 359
Grid Score: -60.092117
Grid_vdw: -62.332535
Grid_es: 2.240418
Int_energy: 13.718960
These files should be copied to local computers if we want to use Chimera to visually check the results. In Seawulf, move to 06.virtual-screen/ directory,
scp –r username@herbie.mathlab.sunysb.edu:~/AMS536/dock_tutorial/
In herbie, open the receptor 1HVR.receptor.mol2 and reference ligand 1HVR.ligand.mol2 and then how the ligands fitting the receptor can be checked by ViewDock function in Chimera. Go to Tools -> Surface/Binding Analysis -> ViewDock and choose 1HVR.output_scored.mol2.In the pup-up window, we can go to Column ->show -> Grid Score to show the grid score.
VII. Running DOCK in Parallel on Seawulf
The Seawulf Cluster is a custom-built 470-processor (235 dual processor nodes and 2 processors per node) Linux Cluster capable of highly parallel processing. It severs as to provide computational resources and expertise for basic scientific research to the faculty and students of Stony Brook University.
Typically, to dock one ligand, one processor on a single node will be used, while for docking multiple ligands, more than one processor can be used in parallel mode. However, the number of processors used should be less than the number of ligands for dock.
Before running DOCK on Seawulf, we need to copy our whole dock-tutorial files from Herbie to Seawulf. Log into the Seawulf through Herbie, and then type,
scp -r username@herbie.mathlab.sunysb.edu:~/AMS536/dock-tutorial/ ~/AMS536/
First, making a file named qsub.csh with the following context,
#!/bin/csh #PBS -l nodes=2:ppn=2 #PBS -l walltime=10:00:00 #PBS -N 1HVR.vs #PBS -o 1HVR.output #PBS -j oe #PBS –V cd /nfs/user03/username/AMS536/dock-tutorial/06.virtual-screen mpirun -np 4 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i virtual_screen.in -o virtual_screen.out
Explanation of the commands,
#!/bin/csh: Execute script with tcsh
#PBS -l nodes=2:ppn=2: Use 2 nodes, and 2 processors per node, so 4 processors
#PBS -l walltime=10:00:00: Allow 10 hours for your job run
#PBS -N 1HVR.vs: Name of your job
#PBS -o 1HVR.output: Name of your output file
#PBS -j oe: Combine the output and error streams into a single output file
#PBS –V: Show more information about what is happening for the users
cd /nfs/user03/username/AMS536/dock-tutorial/06.virtual-screen: Change to your home directory and folder with dock files
mpirun -np 4 /nfs/user03/wjallen/local/dock6/bin/dock6.mpi -i virtual_screen.in -o virtual_screen.out: This line specifies path to dock executable and provide input and output filenames. Notice that in order to run DOCK in parallel (on 4 processors here), we need to use dock6.mpi instead of dock6. Message passing interface ( MPI) is basically a program that allows programs like DOCK to run in parallel.
Then, we can run the dock virtual screen on Seawulf by submitting the DOCK experiment to the Seawulf queue,
qsub virtual_screen.in
You can use the command qstat to show the status of pbs batch jobs,
qstat #the whole list of submitted jobs in queue qstat -u username #the whole list of jobs in queue you submitted
For more information, see PBS Queue.
VIII. Frequently Encountered Problems
- You should always be clear about what the input file and output file of one program are and what format they are.
- Sometimes, it's easy to make mistakes if you are confused about your current location, or "path", so make sure you know that all the time.
- The most common problem experienced in UNIX is having the wrong file path. ALWAYS CHECK TO MAKE SURE THAT YOUR FILE PATH IS CORRECT This can be facilitated with the Tab button which will auto complete what you have started to type.
- Make sure you are opening the correct files for each visualization step (e.g. opening the receptor file with the ligand still there may make analyzing your docking results a bit more challenging).
- Use the auto-fill (tab) function but double check to make sure the correct file was auto-filled (e.g. if you type 1H then hit tab you might get 1HVR_selected_spheres but the file you want actually is 1HVR_selected_spheres.sph).
- For users with no UNIX experience and limited programming experience - it's a good idea to run through a couple tutorials before you start using UNIX. Even if you type in the commands without fully understanding their implications, become familiar with the basic commands and shortcuts. It will help a lot when you actually start using UNIX!
- If you are not so familiar with UNIX environment, it is better for you to practice the tutorial with basic commands: http://www.ee.surrey.ac.uk/Teaching/Unix/
- When you are trying a command, but have a typo (so this command will not work), we can always go back to the last command by press UP arrow button go back to the command that just typed and modify that command.
- Insert button can allows us to retype the command instead of rewrite it.
- Example for writing text, writing a command, and importing an image:
Write some text here..
command or input file