Difference between revisions of "2016 DOCK tutorial with Beta Trypsin"
Stonybrook (talk | contribs) (→Grid computing) |
(→Grid computing) |
||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 50: | Line 50: | ||
Using vi command to open 1BJU.pdb, then deleting bottom and top lines which do not have ATOM or ATPATM in its start. | Using vi command to open 1BJU.pdb, then deleting bottom and top lines which do not have ATOM or ATPATM in its start. | ||
Then changing all ATPATM to ATOM using command: | Then changing all ATPATM to ATOM using command: | ||
| − | %s /ATPATM/ATOM /gc | + | :%s /ATPATM/ATOM /gc |
In addition, changing all GP6 to LIG B using command: | In addition, changing all GP6 to LIG B using command: | ||
| − | %s /GP6 /LIG B /gc | + | :%s /GP6 /LIG B /gc |
Saving this as raw_1BJU.pdb in 00.files. | Saving this as raw_1BJU.pdb in 00.files. | ||
| Line 159: | Line 159: | ||
Since the electronic interactions between two objects in space can be decomposed into pairwise interactions, it is highly efficient to fully calculate a proteins electric contribution to binding energy before DOCKing a ligand. | Since the electronic interactions between two objects in space can be decomposed into pairwise interactions, it is highly efficient to fully calculate a proteins electric contribution to binding energy before DOCKing a ligand. | ||
switch to the 03.box-grid/ directory | switch to the 03.box-grid/ directory | ||
| − | cd 03.box-grid/ | + | cd 03.box-grid/ |
create an input file named showbox.in | create an input file named showbox.in | ||
| − | vim showbox.in | + | |
| + | vim showbox.in | ||
enter the following information | enter the following information | ||
Y # Do you want to calculate a box? (YES) | Y # Do you want to calculate a box? (YES) | ||
| Line 169: | Line 170: | ||
1BJU.box.pdb # What do we want to name the output file? | 1BJU.box.pdb # What do we want to name the output file? | ||
save the file, then enter the following command to generate the box | save the file, then enter the following command to generate the box | ||
| − | showbox | + | showbox < showbox.in |
The box can be visualized in Chimera by loading the output_spheres_selected.pdb file, then loading the 1BJU.box.pdb file | The box can be visualized in Chimera by loading the output_spheres_selected.pdb file, then loading the 1BJU.box.pdb file | ||
| Line 178: | Line 179: | ||
Create the input file for grid. Enter the command | Create the input file for grid. Enter the command | ||
| − | vim grid.in | + | vim grid.in |
Run grid with the command | Run grid with the command | ||
| − | grid - grid.in | + | grid -i grid.in |
This allows you to go back and fix the input file in case there is a mistake when answer the questions prompted by grid. | This allows you to go back and fix the input file in case there is a mistake when answer the questions prompted by grid. | ||
| + | grid.in | ||
| − | + | Parameter Value Description | |
| − | Parameter Value Description | ||
| − | compute_grids yes we want to compute grids | + | compute_grids yes we want to compute grids |
| − | grid_spacing 0.4 resolution of grid energetic contributions (Angstroms). The smaller the value, the more accurate the calculation | + | grid_spacing 0.4 resolution of grid energetic contributions (Angstroms). The smaller the value, the more accurate the calculation |
| − | output_molecule no we don't want to rewrite ligand output file | + | output_molecule no we don't want to rewrite ligand output file |
| − | contact_score no we don't want to deal with contact score | + | contact_score no we don't want to deal with contact score |
| − | energy_score yes we want to compute energetic interaction based on force field | + | energy_score yes we want to compute energetic interaction based on force field |
| − | energy_cutoff_distance 9999 what distance should we stop computing atomic interactions in angstroms | + | energy_cutoff_distance 9999 what distance should we stop computing atomic interactions in angstroms |
| − | atom_model a corresponds to all atom model rather than united atom | + | atom_model a corresponds to all atom model rather than united atom |
| − | attractive_exponent 6 see Lennard-Jones attraction term | + | attractive_exponent 6 see Lennard-Jones attraction term |
| − | repulsive_exponent 12 see Lennard-Jones repulsion term | + | repulsive_exponent 12 see Lennard-Jones repulsion term |
| − | distance_dielectric yes linear decrease in electric force | + | distance_dielectric yes linear decrease in electric force |
| − | dielectric_factor 4 how much will electric force decrease based on atomic distance in angstroms | + | dielectric_factor 4 how much will electric force decrease based on atomic distance in angstroms |
| − | bump_filter yes | + | bump_filter yes |
| − | bump_overlap 0.75 | + | bump_overlap 0.75 |
| − | receptor_file ../01.dockprep/1BJU.rec.mol2 | + | receptor_file ../01.dockprep/1BJU.rec.mol2 |
| − | box_file 4TKG.box.pdb | + | box_file 4TKG.box.pdb |
| − | vdw_definition_file ../zzz.parameters/vdw_AMBER_parm99.defn characteristic van der Waal radii parameter file | + | vdw_definition_file ../zzz.parameters/vdw_AMBER_parm99.defn characteristic van der Waal radii parameter file |
| − | score_grid_prefix grid ONLY THE PREFIX OF THE GRID FILE NAME | + | score_grid_prefix grid ONLY THE PREFIX OF THE GRID FILE NAME |
==V. Docking a Single Molecule for Pose Reproduction== | ==V. Docking a Single Molecule for Pose Reproduction== | ||
| Line 737: | Line 738: | ||
==VIII. Frequently Encountered Problems== | ==VIII. Frequently Encountered Problems== | ||
EVERYONE | EVERYONE | ||
| + | |||
| + | You may see that your vdw scores are zero - this is highly unlikely to actually be the case. If you are using an older version of Chimera and used the "dockprep" button to add charges and hydrogens to your receptor, the charges were likely not saved. You must add the charges and hydrogens manually. | ||
| + | |||
| + | When submitting the qsub script, make sure that there are no spaces after the " \ " on the mpirun line - the job will not run in parallel if there are spaces after the backslash. | ||
Latest revision as of 17:53, 30 January 2017
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2016, using DOCK v6.8.
Contents
I. Introduction
DOCK
DOCK is a molecular docking program used in drug discovery. It was developed by Irwin D. Kuntz, Jr. and colleagues at UCSF (see UCSF DOCK). This program, given a protein binding site and a small molecule, tries to predict the correct binding mode of the small molecule in the binding site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug. DOCK works well as a screening procedure for generating leads, but is not currently as useful for optimization of those leads.
DOCK 6 uses an incremental construction algorithm called anchor and grow. It is described by a three-step process:
- Rigid portion of ligand (anchor) is docked by geometric methods.
- Non-rigid segments added in layers; energy minimized.
- The resulting configurations are 'pruned' and energy re-minimized, yielding the docked configurations.
Beta Trypsin
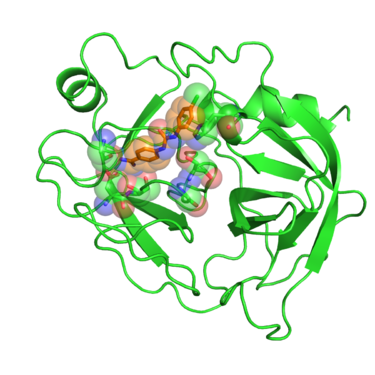
Beta trypsin is a serine protease involved in degrading peptides and proteins. The development of inhibitors of trypsin-like serine proteases has been justified because of the wide processes these enzymes regulate, among them blood coagulation, complement activation, digestion, reproduction and phagocytosis. Early developments of inhibitors for these enzymes were based on the benzamidine chemical structure. Crystal structures of these benzamidine based inhibitors in complex with beta trypsin gave showed the key resides interactions with the inhibitors and pointed out the importance of the "oxyanion hole" within the catalytic site. This gave new insights of the reaction mechanism of the enzyme and revealed what interactions are required to develop inhibitors with increased potency. [1]
In this tutorial we will use PDB website 1BJU deposited crystal structure of beta-trypsin in complex with inhibitor GP6.
Organizing Directories
While performing docking, it is convenient to adopt a standard directory structure / naming scheme, so that files are easy to find / identify.For this tutorial, we will use something similar to the following:
~username/AMS536-Spring2016/dock-tutorial/00.files/
/01.dockprep/
/02.surface-spheres/
/03.box-grid/
/04.dock/
/05.large-virtual-screen/
/06.virtual-screen/
/07.footprint/
/08.print_fps
|
In addition, most of the important files that are derived from the original crystal structure will be given a prefix that is thsame as the PDB code, '1BJU'.The following sections in this tutorial will adhere to this directory structure/naming scheme.
II. Preparing the Receptor and Ligand
Download the PDB File (1BJU)
Go to the PDB website to download 1BJU.pdb file(PDB code: 1BJU). Using command or WinSCP to transfer 1BJU.pdb to 00.files. If you are in 00.files now, the command is:
cp ~/Downloads/1BJU.pdb .
Edit the PDB File (1BJU)
Using vi command to open 1BJU.pdb, then deleting bottom and top lines which do not have ATOM or ATPATM in its start. Then changing all ATPATM to ATOM using command:
:%s /ATPATM/ATOM /gc
In addition, changing all GP6 to LIG B using command:
:%s /GP6 /LIG B /gc
Saving this as raw_1BJU.pdb in 00.files.
Prepare ligand and receptor files for dock
Create dockprep file
Open raw_1BJU.pdb file in chimera, in Tools, find structure editing, then click AddH to add hydrogen in this system.
Also in structure editing, click Add charge, then, changing AMBER ff14SB to AMBER ff99SB and changing net charge to +1.
Saving it as 1BJU.dockprep.mol2 in 01.dockprep.
Create receptor file
Open 1BJU.dockprep.mol2 in chimera, then click select to choose ligand, click action in Atoms/Bonds click delete.
Saving this molecule as 1BJU.rec.mol2 in 01.dockprep.
Create ligand file
Open 1BJU.dockprep.mol2 in chimera, using the same method delete the protein molecule.
Saving it as 1BJU.lig.mol2 in 01.dockprep.
Create no hydrogen receptor file
Open 1BJU.rec.mol2 in chimera, click select, then choosing chemistry H to select hydrogen in protein, Using action button to delete H.
Saving it as 1BJU.rec.noH.pdb in 01.dockprep.
III. Generating Receptor Surface and Spheres
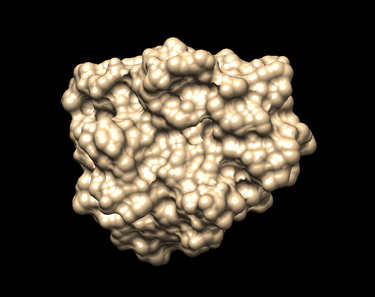
Generating the Receptor Surface
Create the directory where the following steps will be done:
mkdir 02.surface-sphere cd 02.surface-sphere
Open Chimera in the terminal to view the receptor surface:
Chimera
In Chimera complete the following steps to open the receptor file in the 01.dockprep directory:
File -> Open -> 1BJU.rec.noH.pdb
To show the surface receptor:
Action -> Surface -> Show (The surface should look like the picture shown below.)
To save a DMS file as 1BJU.rec.dms in 02.surface-sphere directory:
Tools -> Structure editing -> Write DMS
Creating Spheres
Using the Sphgen program complete following steps:
1. Create an input file name INSPH to be read by Sphgen:
vi INSPH
1BJU.rec.dms #specifies the input file R #spheres generated will be outside of teh receptor surface X #specifies that all points won the receptor will be used 0.0 #distance in angstroms (avoids steric clashes) 4.0 #max surface radius of the spheres in angstroms 1.4 #min surface radius of the spheres in angstroms 1BJU.rec.sph #the specified outfile containing all generated spheres
2. Run the Sphgen using the input file INSPH with the command:
sphgen -i INSPH -o OUTSPH
-i is the flag that specifies the input file INSPH INSPH is int eh specified input file -o is the flag to specifiy the output file OUTSPH is the file containing the generated spheres
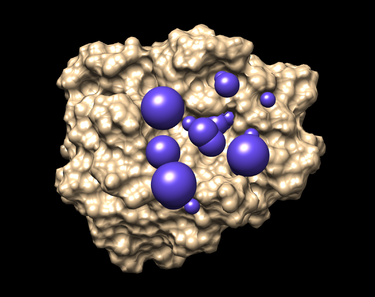
3. Visualization generated spheres with Chimera:
- Launch Chimera in termal,
- Choose File -> Open, choose 1BJU.rec.mol2
- choose File -> Open, choose 1BJU.rec.sph
You should have an image like this:
Selecting Spheres
To select spheres of interest run the program sphere_selector in the terminal at 8.0 angstrom radius of the target molecule or known binding site:
sphere_selector 1BJU.rec.sph ../01.dockprep/1BJU.lig.mol2 8.0
The output file generated will be selected_spheres.sph
To visualize the spheres using Chimera as previously done:
- Launch Chimera, choose File -> Open, choose 1BJU.rec.noH.pdb
- File -> Open, choose output_spheres_selected.pdb
- Select -> Residue -> SPH
- Actions -> Atoms/Bonds -> sphere
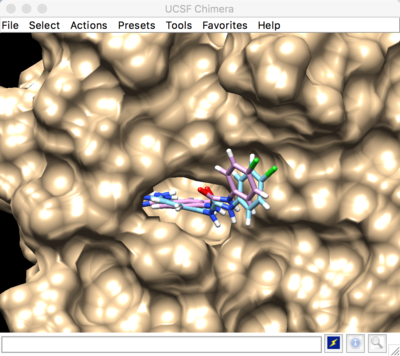
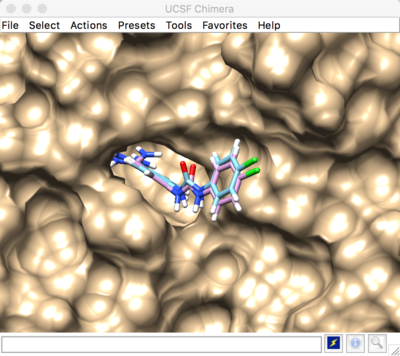
The selected spheres with the receptor surface should look similar to that as seen below:
IV. Generating Box and Grid
Monaf Awwa
Box Generation
Since the electronic interactions between two objects in space can be decomposed into pairwise interactions, it is highly efficient to fully calculate a proteins electric contribution to binding energy before DOCKing a ligand. switch to the 03.box-grid/ directory
cd 03.box-grid/
create an input file named showbox.in
vim showbox.in
enter the following information
Y # Do you want to calculate a box? (YES) 8.0 # How long should the box length be? ../02.surface-spheres/selected_spheres.sph # which sphere file is our input? 1 # How many clusters for the file? 1BJU.box.pdb # What do we want to name the output file?
save the file, then enter the following command to generate the box
showbox < showbox.in
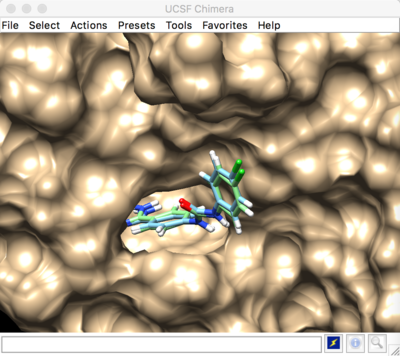
The box can be visualized in Chimera by loading the output_spheres_selected.pdb file, then loading the 1BJU.box.pdb file
Grid computing
Next, we will calculate the grid energies
Create the input file for grid. Enter the command
vim grid.in
Run grid with the command
grid -i grid.in
This allows you to go back and fix the input file in case there is a mistake when answer the questions prompted by grid.
grid.in
Parameter Value Description
compute_grids yes we want to compute grids
grid_spacing 0.4 resolution of grid energetic contributions (Angstroms). The smaller the value, the more accurate the calculation
output_molecule no we don't want to rewrite ligand output file
contact_score no we don't want to deal with contact score
energy_score yes we want to compute energetic interaction based on force field
energy_cutoff_distance 9999 what distance should we stop computing atomic interactions in angstroms
atom_model a corresponds to all atom model rather than united atom
attractive_exponent 6 see Lennard-Jones attraction term
repulsive_exponent 12 see Lennard-Jones repulsion term
distance_dielectric yes linear decrease in electric force
dielectric_factor 4 how much will electric force decrease based on atomic distance in angstroms
bump_filter yes
bump_overlap 0.75
receptor_file ../01.dockprep/1BJU.rec.mol2
box_file 4TKG.box.pdb
vdw_definition_file ../zzz.parameters/vdw_AMBER_parm99.defn characteristic van der Waal radii parameter file
score_grid_prefix grid ONLY THE PREFIX OF THE GRID FILE NAME
V. Docking a Single Molecule for Pose Reproduction
Minimization
One can use the DOCK software to perform a energy minimization of a ligand in the receptor's binding site.
Create an empty file (min.in) and run dock6:
touch min.in dock6 -i min.in
The dock program will start and it will begin to ask you a series of questions. The answers will be saved to the min.in file, so next time you don't have to go through all the questions again. You can just edit the min.in file directly.
Dock Minimization Input file: min.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../zzz.parameters/vdw.defn flex_defn_file ../zzz.parameters/flex.defn flex_drive_file ../zzz.parameters/flex_drive.tbl ligand_outfile_prefix 1BJU.min write_orientations no num_scored_conformers 1 rank_ligands no
Note that "conformer_search_type" is set to "rigid".
Note that "simplex_restraint_min" is set to "yes". This is a flag to perform RMSD tethering, which restrains the conformer sampling to not move too far away from the initial docking site.
The output file prefix is "1BJU.min". The output file "1BJU.min_scored.mol2" is saved in the directory where you ran dock6 (04.dock). This file contains the predicted pose coordinates, internal energy, grid score, and RMSD values.
To view your results, you can open the 1BJU.min_scored.mol2 file in CHIMERA and compare it to your original ligand file in the 01.dockprep directory.
Minimization result:
The energy minimized pose has an RMSD of 0.3177, meaning that it was minimally moved from it's original experimental ligand pose (1BJU.lig.mol2).
Rigid docking
Rigid Ligand Docking is typically used to check the validity of docking program, user-defined parameters, and help identify any mistakes in the receptor and ligand prep files. Here, we dock the ligand back to the ligand-free receptor. The pose predicted by DOCK should be very close to the crystal structure of the ligand-receptor complex.
The following should be performed in the 04.dock directory.
Create an empty file (rigid_r.in) and run DOCK6:
touch rigid_r.in dock6 -i rigid_r.in
Rigid docking input file: rigid_r.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphere/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../zzz.parameters/vdw.defn flex_defn_file ../zzz.parameters/flex.defn flex_drive_file ../zzz.parameters/flex_drive.tbl ligand_outfile_prefix 1BJU.rigid_restraint write_orientations no num_scored_conformers 40 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Note that "conformer_search_type" is set to "rigid".
Note that simplex_restraint_min is set to "yes". This is a flag to perform RMSD tethering, which restrains the conformer sampling to not move too far away from the initial docking site.
The output file prefix is "1BJU.rigid_restraint". The output file "1BJU.rigid_restraint_scored.mol2" is saved in the directory where you ran dock6 (04.dock). This file contains the ligand coordinates, energy score, and RMSD values.
To view your results, you can open the pdb files of the receptor and ligand in CHIMERA from your 01.dockprep directory. The open your rigid docking results using viewdock:
Tools > surface/binding analysis > viewdock > 1BJU.rigid_restraint_scored.mol2.
You can rank the conformers based on internal energy, grid score, or RMSD.
Best scored rigid docking result:
The predicted pose and the experimental pose have an RMSD less than 2.000, meaning that we were able to reproduce the experimental ligand pose (crystal structure).
Flexible Docking
Flexible docking samples internal degrees of freedom.
The following should be performed in the 04.dock directory.
Create an empty file (flex.in) and run DOCK6:
touch flex.in dock6 -i flex.in
Flexible docking input file: flex.in
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphere/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file ../zzz.parameters/vdw.defn flex_defn_file ../zzz.parameters/flex.defn flex_drive_file ../zzz.parameters/flex_drive.tbl ligand_outfile_prefix 1BJU.flex write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Note that "conformer_search_type" is set to "flex".
The output file prefix is "1BJU.flex" and the output file is saved as "1BJU.flex_scored.mol2" in the 04.dock directory. The file "1BJU.flex_scored.mol2" contains the ligand coordinates, energy score, and RMSD values. To view your results, you can open the pdb files of the receptor and ligand in CHIMERA from your 01.dockprep directory. Then open your flexible docking results using viewdock:
Tools > surface/binding analysis > viewdock > 1BJU.flex_scored.mol2.
You can rank the conformers based on internal energy, grid score, or RMSD.
Best scored flexible docking result:
The predicted pose and the experimental pose have an RMSD less than 2.000, meaning that we were able to reproduce the experimental ligand pose (crystal structure). If your top scored conformer has a high RMSD, this is considered a DOCKing failure. In the event of a DOCKing failure, consider setting "simplex_random_seed" to a different value. This is a random number generator that determines the starting point for conformer sampling. Alternatively, you could increase sampling.
VI. Virtual Screening
Katie Maffucci
Virtual screening uses libraries of small molecules to identify novel chemical structures that are most likely to bind a drug target. The search is narrowed to a few "hit" compounds that can be synthesized or purchased and tested against the target. Structure-based virtual screening is computationally demanding, and requires a parallel computing infrastructure to handle the task. LIRED is a Linux-based computing cluster well suited for this purpose.
DOCK Preparation
To set up the virtual screen, make a directory using the command: "mkdir 05.largevirtualscreen" Print your working directory using the "pwd" command and copy this dock directory into your LIRED account: "scp -r /gfps/home/username/AMS536/docktutorial/05.largevirtualscreen ."
QSUB File
To submit your job to the cluster, a qsub c chell script must be made so that the job can be executed. Its contents should be as follows:
#! /bin/tcsh #PBS -l nodes=2:ppn=2 #PBS -l walltime=24:00:00 #PBS -o zzz.qsub.out #PBS -e zzz.qsub.err #PBS -N large_vs #PBS -M your.name@stonybrook.edu #PBS -V cd /gfps/home/username/AMS536/docktutorial/ /gpfs/software/intel_2016_update1/impi/5.1.2.150/bin64/mpirun np 4 \ /home/tmcgee/local/AMS_536/dock6/bin/dock6.mpi \
Where -l = resources required for the job, -o = defines output,-e = defines error, ppn = processors per node, -N = name of job, walltime = how long the job can run, and -M = to set up an email alert when the job is complete
Input File
An input file must also be made. "vi large_vs.in":
ligand_atom_file yourligandmol2file.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd no
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file ../02.surface-spheres/selected_spheres.sph
max_orientations 1000
critical_points no
chemical_matching no
use_ligand_spheres no
use_internal_energy yes
internal_energy_rep_exp 12
flexible_ligand yes
user_specified_anchor no
limit_max_anchors no
min_anchor_size 5
pruning_use_clustering yes
pruning_max_orients 1000
pruning_clustering_cutoff 100
pruning_conformer_score_cutoff 100
use_clash_overlap no
write_growth_tree no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../03.box-grid/grid
multigrid_score_secondary no
dock3.5_score_secondary no
continuous_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_anchor_max_iterations 500
simplex_grow_max_iterations 500
simplex_grow_tors_premin_iterations 0
simplex_random_seed 0
simplex_restraint_min no
atom_model all
vdw_defn_file ../docktutorial/zzz.parameters/vdw_AMBER_parm99.defn
flex_defn_file ../docktutorial/zzz.parameters/flex.defn
flex_drive_file ../docktutorial/zzz.parameters/flex_drive.tbl
ligand_outfile_prefix large_vs
write_orientations no
num_scored_conformers 1
rank_ligands no
Post Processing
After the virtual screen, the top hits and poses will be rank ordered according to the scoring function(s) chosen in previous input files. Depending on the size of the database, the top 1% of molecules can still number in the hundreds. To further refine the screen, cartesian minimization should be performed. To do this, create an input file 1BJU.cart_min.ligs.in:
conformer_search_type rigid
use_internal_energy yes
internal_energy_rep_exp 12
ligand_atom_file ../06.large-virtual-screen/1BJU.output_scored.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd no
use_database_filter no
orient_ligand no
automated_matching no
critical_points no
chemical_matching no
use_ligand_spheres no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary no
grid_score_secondary no
multigrid_score_primary no
multigrid_score_secondary no
dock3.5_score_primary no
dock3.5_score_secondary no
continuous_score_primary yes
continuous_score_secondary no
cont_score_rec_filename ../01.dockprep/1BJU.rec.mol2
cont_score_att_exp 6
cont_score_rep_exp 12
cont_score_rep_rad_scale 1
cont_score_use_dist_dep_dielectric yes
cont_score_dielectric 4.0
cont_score_vdw_scale 1
cont_score_es_scale 1
footprint_similarity_score_secondary no
ph4_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
simplex_max_iterations 1000
simplex_tors_premin_iterations 0
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_random_seed 0
simplex_restraint_min yes
simplex_coefficient_restraint 10.0
atom_model all
vdw_defn_file ../zzz.parameters/vdw.defn
flex_defn_file ../zzz.parameters/flex.defn
flex_drive_file ../zzz.parameters/flex_drive.tbl
ligand_outfile_prefix 1BJU.cartmin
write_orientations no
num_scored_conformers 1
rank_ligands no
You'll need to make another minimization input file with similar contents:
conformer_search_type rigid
use_internal_energy yes
internal_energy_rep_exp 12
ligand_atom_file ../01.dockprep/1BJU.lig.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd no
use_database_filter no
orient_ligand no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary no
grid_score_secondary no
multigrid_score_primary no
multigrid_score_secondary no
dock3.5_score_primary no
dock3.5_score_secondary no
continuous_score_primary yes
continuous_score_secondary no
cont_score_rec_filename ../01.dockprep/1BJU.rec.mol2
cont_score_att_exp 6
cont_score_rep_exp 12
cont_score_rep_rad_scale 1
cont_score_use_dist_dep_dielectric yes
cont_score_dielectric 4.0
cont_score_vdw_scale 1
cont_score_es_scale 1
footprint_similarity_score_secondary no
ph4_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
simplex_max_iterations 1000
simplex_tors_premin_iterations 0
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_random_seed 0
simplex_restraint_min yes
simplex_coefficient_restraint 10.0
atom_model all
vdw_defn_file ../zzz.parameters/vdw.defn
flex_defn_file ../zzz.parameters/flex.defn
flex_drive_file ../zzz.parameters/flex_drive.tbl
ligand_outfile_prefix 1BJU.cartmin_ref
write_orientations no
num_scored_conformers 1
rank_ligands no
To execute the job, you'll need to make a cartminligs.qsub.tcsh script similar to the previous one:
#!/bin/tcsh #PBS -l walltime=09:00:00 #PBS -l nodes=1:ppn=24 #PBS -q medium #PBS -N 1BJU #PBS -V #PBS -M youremail@stonybrook.edu #PBS -j oe cd /gpfs/home/username/AMS536/docktutorial/07.cartesian_min /gpfs/software/intel_2016_update1/impi/5.1.2.150/bin64/mpirun -np 24 \ /home/tmcgee/local/AMS_536/dock6/bin/dock6.mpi \ -i 1BJU.cart_min_ligs.in \ -o 1BJU.cart_min_ligs.out
VIII. Frequently Encountered Problems
EVERYONE
You may see that your vdw scores are zero - this is highly unlikely to actually be the case. If you are using an older version of Chimera and used the "dockprep" button to add charges and hydrogens to your receptor, the charges were likely not saved. You must add the charges and hydrogens manually.
When submitting the qsub script, make sure that there are no spaces after the " \ " on the mpirun line - the job will not run in parallel if there are spaces after the backslash.