Difference between revisions of "2021 Denovo tutorial 3 with PDBID 1S19"
Stonybrook (talk | contribs) (→Fragment Libraries) |
Stonybrook (talk | contribs) (→Fragment Libraries) |
||
| Line 55: | Line 55: | ||
grep -wc MOLECULE *.mol2 | grep -wc MOLECULE *.mol2 | ||
| + | |||
| + | You should get the following output upon using this command: | ||
| + | |||
| + | fraglib_linker.mol2:4 | ||
| + | fraglib_rigid.mol2:0 | ||
| + | fraglib_scaffold.mol2:2 | ||
| + | fraglib_sidechain.mol2:2 | ||
| + | fragment.out_scored.mol2:0 | ||
| + | |||
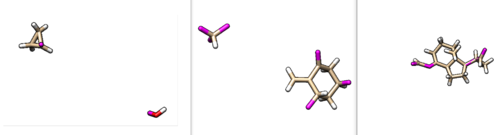
These fragments can also be visualized in Chimera. The results are shown below. | These fragments can also be visualized in Chimera. The results are shown below. | ||
Revision as of 16:40, 8 March 2021
This tutorial continues where we left off from the virtual screening of 1S19, a structure of the vitamin D nuclear receptor bound to the side chain analogue calcipotriol.
Contents
Focused Denovo Design with PDB 1S19
In this section, we will generate new ligand structures using a fragment library. It will require the use of some of the output files from the Virtual Screen tutorial (found here).
Fragment Libraries
We will use a focused fragment library file based on the original ligand. To begin, we will create a new directory for this.
mkdir fraglib
Create a new file called fragment.in with the following inputs.
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001_structure/1s19_ligand_dockprep.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix fragment.out write_orientations no num_scored_conformers 1 rank_ligands no
To run this, type:
dock6 -i fragment.in -o fragment.out
Once this has ran, the following files should have been generated: fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_sidechain.mol2, fraglib_torenv.dat. To check the number of fragments in each file, you can type:
grep -wc MOLECULE *.mol2
You should get the following output upon using this command:
fraglib_linker.mol2:4 fraglib_rigid.mol2:0 fraglib_scaffold.mol2:2 fraglib_sidechain.mol2:2 fragment.out_scored.mol2:0
These fragments can also be visualized in Chimera. The results are shown below.