Difference between revisions of "2023 AMBER tutorial 1 with PDBID 4S0V"
Stonybrook (talk | contribs) (→Complex Equilibration) |
Stonybrook (talk | contribs) (→Complex Equilibration) |
||
| Line 219: | Line 219: | ||
=Complex Equilibration= | =Complex Equilibration= | ||
| − | Before we can run our structure through the AMBER program we need to minimize it and make sure it's at equilibrium. | + | Before we can run our structure through the AMBER program we need to minimize it and make sure it's at equilibrium. These commands will again be done on the command line so please cd into your 004.equil directory. |
| − | + | Nine input files are required for system equilibration and AMBER will process them in order. You can imagine these steps as 'heat-treating' your complex - you apply heat to the system and then let it relax - and repeat until the system is at rest. Each of these nine files is one step in this process. For each step there are two files: | |
| − | |||
| − | |||
| − | |||
min.mdin - will minimize only hydrogen atoms | min.mdin - will minimize only hydrogen atoms | ||
equil.mdin - will minimize system including non-hydrogen atoms, depending upon the restraint mask for atoms involved | equil.mdin - will minimize system including non-hydrogen atoms, depending upon the restraint mask for atoms involved | ||
| − | + | These input files will also direct AMBER on which atoms to restrain during equilibration. There a specific line in each file: | |
| + | restraintmask | ||
| + | |||
| + | after that command, if you see a "!", that is telling AMBER to exclude any atoms that follow; if you see "@H=", that is telling AMBER to restrain all the hydrogen atoms. In the first 7 input files we want to restrain all residues in our complex, including the ligand. For the last two steps we will only restrain the protein and not the ligand. | ||
| − | + | There is also a line: | |
| + | restraint_wt | ||
| − | What are restraint weights? Check the AMBER handbook for a better description, but in brief, | + | What are restraint weights? Check the AMBER handbook for a better description, but in brief, this is a constant similar to a spring constant used in Hooke's law. For the first files we use a larger restraint_wt and a lower constant for the final files. Before we can create any input files we need to know how many residues are in our complex. Do this by looking at at the 4s0v.gas.complex.pdb file created in the previous step and looking at the last line: |
[[File: complexpdb.png|center]] | [[File: complexpdb.png|center]] | ||
| Line 525: | Line 526: | ||
sbatch 4s0v.equil.slurm | sbatch 4s0v.equil.slurm | ||
| − | Once the program finishes you will see | + | Once the program finishes you will see multiple new files including a file named logfile. Check to file for errors. If there are none, you can move onto the next step. You should also check the .trj files from each step to make sure that the system appears rational. Below is the equillibrated system following 09.equil.mdin |
[[File: 4S0V_MIN.png|center||500px]] | [[File: 4S0V_MIN.png|center||500px]] | ||
Revision as of 17:29, 20 March 2023
Contents
Introduction
AMBER is a molecular dynamics program that can be run on your protein/ligand complex to ensure that the interactions between the two structures are stable. DOCK shows us how the two interact with each other at one point in time. AMBER looks at those interactions over time to ensure that forces will not occur which will push the ligand out of the binding site as the complex naturally moves. This tutorial will again be working with PDB #4s0v
Setting Up Your Environment
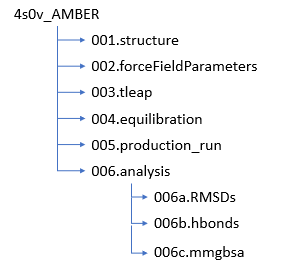
Just as with DOCK you should set up for directory structure at this point to keep everything organized and easy to find. We will be creating a new structure which looks like:
Structure
Before starting the analysis it's best to download a new protein/ligand complex from the PDB and isolate both the protein and ligand structures.
Tools -> General Controls -> Command Line
on the command line, type:
open 4sov
then:
Select -> Structure -> protein
which will select the receptor, and then go to
Select -> Invert (all models)
to select everything other than the receptor in the file. Then, go to
Actions -> Atoms/Bonds -> delete
which will isolate the receptor.
For the protein file, it’s important to model any missing loops before running AMBER on the complex. You might not have done this for the docking tutorial because the loops were far enough from the binding site to not matter but for this step it needs to be done. After adding in any missing loops with
Tools -> Structure Editing -> Model/Refine Loops
Go to
File -> Save Mol2...
and save as 4s0v_nolig_noH_nocharge.mol2. This will save a mol2 of just the protein, without hydrogens or charges added.
Alternatively, you can follow the steps in the *[1] tutorial to do this. The inputs we need are the isolated protein with NO hydrogens and NO charges; and the ligand with hydrogens and charges. In other words, once you isolate the protein structure in Chimera, save it with a filename such as, 4s0v_protein_for_AMBER.pdb.
Then isolate the ligand structure, add hydrogens and re-do whatever protonation changes you made in the *[2] tutorial.
For our purposes in this tutorial, we will leave the ligand as a .pdb for parametrization, since defining the connectivity of the oxygens that we removed hydrogens from can be tricky. However, if you are following this for your own ligands, you can explicitly define the charges etc if you are confident.
We save this ligand file (with hydrogens) as: 4s0v_ligand_only.pdb
Once these two files have been generated, scp them over to the 001.structure directory on Seawulf.
Force Field Parameters
AMBER needs force field parameters to run correctly. This only needs to be done for the ligand.
Generate a 002.parameters directory and cd into it.
To generate ligand parameters execute the following slurm script:
#!/bin/bash # #SBATCH --job-name=4s0v_AMBER_parameters #SBATCH --output=parameters_output.txt #SBATCH --ntasks-per-node=24 #SBATCH --nodes=6 #SBATCH --time=48:00:00 #SBATCH -p long-24core module load amber/16 antechamber -i ../001.structure/4s0v_ligand_only.pdb -fi pdb -o 4s0v_ligand_antechamber.mol2 -fo mol2 -at gaff2 -c bcc -rn LIG -nc 0
The important parameter in the above command is the -nc option at the end. This is telling antechamber what the total charge is on your ligand. This number needs to be the same as the number used in the previous step when you added charges to the structure in Chimera. In our case, the total charge on the ligand should be 0.
Once this has completed running you will see multiple new files in your directory including, specifically:
4s0v_ligand_antechamber.mol2 ANTECHAMBER_AC.AC0 ANTECHAMBER_AM1BCC_PRE.AC ANTECHAMBER_BOND_TYPE.AC0 sqm.in sqm.pdb ANTECHAMBER_AC.AC ANTECHAMBER_AM1BCC.AC ANTECHAMBER_BOND_TYPE.AC ATOMTYPE.INF sqm.out
4s0v_ligand_antechamber.mol2 is the file with the parameters we just generated.
Now that we have defined parameters for the ligand, we need to modify our force field parameters slightly. Run the command:
parmchk2 -i 4s0v_ligand_antechamber.mol2 -f mol2 -o 4s0v_ligand.am1bcc.frcmod
If you get an error when running this line, try typing:
module load amber/16
and then running the command again. Once it's done running you will see the 4s0v_ligand.am1bcc.frcmod file in your directory.
TLeap Implemenation
The goal of molecular dynamics simulations is to model the behavior of molecular systems and observe the behavior and interactions between constituents in those systems. Here, we investigate the interactions between our ligand and our receptor. Our MD analysis using AMBER will allow is to experimentally model this interactions over a given timeframe, and from these simulations we can estimate the affinity of the receptor for this ligand.
mkdir 003.tleap
We will first generate the gas-phase and solvated systems, and neutralize both systems.
To create the input script (which should be run on the cluster, not on a login node, see below):
nano 4s0v_tleap.in
And then add:
#!/usr/bin/sh
###load protein force field
source leaprc.protein.ff14SB
###load GAFF force field (for our ligand)
source leaprc.gaff
###load TIP3P (water) force field
source leaprc.water.tip3p
###load ions frcmod for the tip3p model
loadamberparams frcmod.ionsjc_tip3p
###needed so we can use igb=8 model
set default PBradii mbondi3
###load protein pdb file
rec=loadpdb ../001.structure/4s0v_built.pdb
#THIS IS WHERE YOU WOULD DEFINE DISULFIDE BONDS
#NUMBERING SHOULD MATCH INPUT PDB FILE
#bond rec.649.SG rec.762.SG
#bond rec.454.SG rec.472.SG
#bond rec.444.SG rec.447.SG
#bond rec.328.SG rec.339.SG
#bond rec.385.SG rec.394.SG
###load ligand frcmod/mol2
loadamberparams ../002.parameters/4s0v_ligand.am1bcc.frcmod
lig=loadmol2 ../002.parameters/4s0v_ligand_antechamber.mol2
###create gase-phase complex
gascomplex= combine {rec lig}
###write gas-phase pdb
savepdb gascomplex 4s0v.gas.complex.pdb
###write gase-phase toplogy and coord files for MMGBSA calc
saveamberparm gascomplex 4s0v.complex.parm7 4s0v.gas.complex.rst7
saveamberparm rec 4s0v.gas.receptor.parm7 4s0v.gas.receptor.rst7
saveamberparm lig 4s0v.gas.ligand.parm7 4s0v.gas.ligand.rst7
###create solvated complex (albeit redundant)
solvcomplex= combine {rec lig}
###solvate the system
solvateoct solvcomplex TIP3PBOX 12.0
###Neutralize system
addions solvcomplex Cl- 0
addions solvcomplex Na+ 0
#write solvated pdb file
savepdb solvcomplex 4s0v.wet.complex.pdb
###check the system
charge solvcomplex
check solvcomplex
###write solvated toplogy and coordinate file
saveamberparm solvcomplex 4s0v.wet.complex.prmtop 4s0v.wet.complex.rst7
quit
This input file should be executed on the cluster, not in a login node. To do so, create and run the following script: 4s0v_tleap.slurm.
#!/bin/bash # #SBATCH --job-name=4s0v_tleap #SBATCH --output=tleap_output.txt #SBATCH --ntasks-per-node=24 #SBATCH --nodes=6 #SBATCH --time=48:00:00 #SBATCH -p long-96core module load amber/16 tleap -f 4s0v_tleap.in
Execute this script with:
sbatch 4s0v_tleap.slurm
When this is done running you will see multiple files in your directory. Specifically:
4s0v.complex.parm7 4s0v.gas.complex.rst7 4s0v.gas.ligand.rst7 4s0v.gas.receptor.rst7 4s0v_tleap.slurm 4s0v.wet.complex.prmtop leap.log 4s0v.gas.complex.pdb 4s0v.gas.ligand.parm7 4s0v.gas.receptor.parm7 4s0v_tleap.in 4s0v.wet.complex.pdb 4s0v.wet.complex.prmtop 4s0v.wet.complex.rst7 tleap_output.txt
Scp 4s0v.wet.complex.rst7 and 4s0v.wet.complex.prmtop to your local computer.
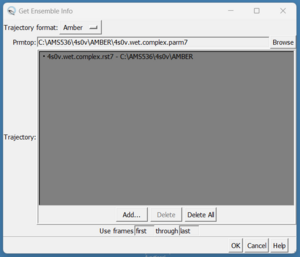
In Chimera, open using:
Tools → MD/Ensemble Analysis → MD Movie
In the prmtop section, select 4s0v.wet.complex.prmtop, and then hit "Add" and select the 4s0v.wet.complex.rst7 file. Your receptor should now open in Chimera.
Everything looks good so we can move onto the next step.
Complex Equilibration
Before we can run our structure through the AMBER program we need to minimize it and make sure it's at equilibrium. These commands will again be done on the command line so please cd into your 004.equil directory.
Nine input files are required for system equilibration and AMBER will process them in order. You can imagine these steps as 'heat-treating' your complex - you apply heat to the system and then let it relax - and repeat until the system is at rest. Each of these nine files is one step in this process. For each step there are two files:
min.mdin - will minimize only hydrogen atoms equil.mdin - will minimize system including non-hydrogen atoms, depending upon the restraint mask for atoms involved
These input files will also direct AMBER on which atoms to restrain during equilibration. There a specific line in each file:
restraintmask
after that command, if you see a "!", that is telling AMBER to exclude any atoms that follow; if you see "@H=", that is telling AMBER to restrain all the hydrogen atoms. In the first 7 input files we want to restrain all residues in our complex, including the ligand. For the last two steps we will only restrain the protein and not the ligand.
There is also a line:
restraint_wt
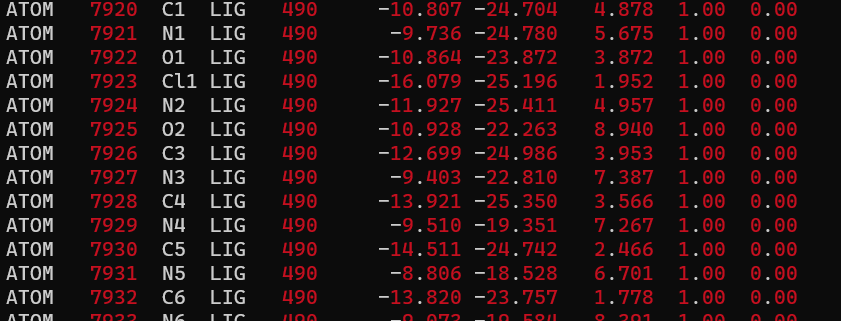
What are restraint weights? Check the AMBER handbook for a better description, but in brief, this is a constant similar to a spring constant used in Hooke's law. For the first files we use a larger restraint_wt and a lower constant for the final files. Before we can create any input files we need to know how many residues are in our complex. Do this by looking at at the 4s0v.gas.complex.pdb file created in the previous step and looking at the last line:
We see that there are 490 residues in our complex. 489 in the protein and one for the ligand.
Create 01.min.mdin
nano 01.min.mdin
Copy the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=5000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
In the above input file you'll see a line:
restraintmask=":1-490@CA,C,N", ! atoms to be restrained.
This is where the number of residues for your system needs to be updated. This change needs to be done for the following input files as well.
Create 02.equil.mdin
nano 02.equil.mdin
Copy the following lines:
MD simualation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create 03.min.mdin
nano 03.min.mdin
Copy the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=2.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create 04.min.mdin
nano 04.min.mdin
Copy the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create 05.min.mdin
nano 05.min.mdin
Copy the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=0.05, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create 06.equil.mdin
nano 06.equil.mdin
Copy the following lines:
MD simualation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=1.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create 07.equil.mdin
nano 07.equil.mdin
Copy the following lines:
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-490 & !@H=", ! atoms to be restrained restraint_wt=0.5, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create 08.equil.mdin
nano 08.equil.mdin
Copy the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-489@CA.C.N", ! atoms to be restrained, only the backbone restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
NOTE: for the above input file we only want to restrain the protein and not the ligand so the '490' needs to be change to '489'.
Create 09.equil.mdin.
nano 09.equil.mdin
Copy the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-489@CA.C.N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
NOTE: for the above input file we only want to restrain the protein and not the ligand so the '490' needs to be change to '489'.
Once you have all nine files in your directory we need to create a slurm script, 4s0v.equil.slurm, to run them:
#!/bin/bash
#
#SBATCH --job-name=4s0v_equilibration
#SBATCH --output=equilibration_output.txt
#SBATCH --ntasks-per-node=24
#SBATCH --nodes=1
#SBATCH --time=48:00:00
#SBATCH -p long-24core
module load amber/16
do_parallel="mpirun -np 80 pmemd.MPI"
prmtop="../003.tleap/4s0v.wet.complex.prmtop"
coords="../003.tleap/4s0v.wet.complex"
MDINPUTS=(01.min 02.equil 03.min 04.min 05.min 06.equil 07.equil 08.equil 09.equil)
for input in ${MDINPUTS[@]}; do
$do_parallel -O -i ${input}.mdin -o ${input}.mdout -p $prmtop -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r ${input}.rst7
coords=$input
done
Submit the above file to Seawulf by typing:
sbatch 4s0v.equil.slurm
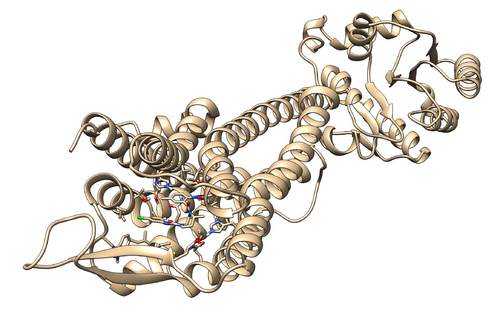
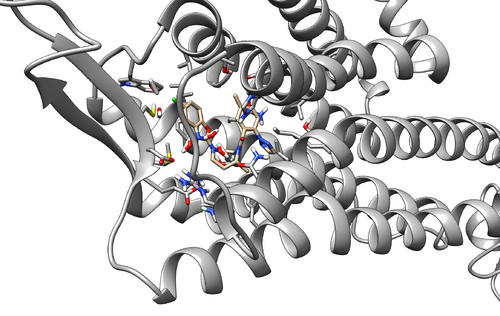
Once the program finishes you will see multiple new files including a file named logfile. Check to file for errors. If there are none, you can move onto the next step. You should also check the .trj files from each step to make sure that the system appears rational. Below is the equillibrated system following 09.equil.mdin
Production Run
Now that our system is set up properly we can move onto an MD production run of the structure. This will again be completed on the command line, please cd into 005.production
mkdir 005.production
Create the input file, 10.prod.min
nano 10.prod.min
And copy the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=5000000, ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=2, ! SHAKE on for bonds with hydrogen dt=0.002, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=2, ! No force evaluation for bonds with hydrogen ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 2500, ! Write to trajectory file every ntwx steps ntpr= 2500, ! Print to mdout every ntpr steps ntwr= 5000000, ! Write a restart file every ntwr steps cut=8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-489@CA,C,N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on
Note for the restraintmask line, use the same number as for the 09.equil.mdin file in the previous step. In our case, this number is 489.
This input file can now be run on Seawulf using the following slurm script:
#!/bin/bash
#
#SBATCH --job-name=4s0v_production
#SBATCH --output=production.txt
#SBATCH --ntasks-per-node=24
#SBATCH --nodes=3
#SBATCH --time=48:00:00
#SBATCH -p long-24core
module load amber/16
do_parallel="/gpfs/software/intel/parallel-studio-xe/2018_3/compilers_and_libraries/linux/mpi/bin64/mpirun -np 80 /gpfs/software/amber/16/intel/cpu/bin/pmemd.MPI"
prmtop="../003.tleap/4s0v.wet.complex.prmtop"
coords="../004.equilibration/09.equil"
MDINPUTS=(10.prod)
for input in ${MDINPUTS[@]}; do
$do_parallel -O -i ${input}.mdin -o ${input}.mdout -p $prmtop -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r ${input}.rst7
coords=$input
done