Difference between revisions of "2024 DOCK tutorial 2 with PDBID 1NDV"
From Rizzo_Lab
Stonybrook (talk | contribs) (→Analyzing the Protein Structure:) |
Stonybrook (talk | contribs) (→Analyzing the Protein Structure:) |
||
| Line 6: | Line 6: | ||
=== Protein === | === Protein === | ||
====Analyzing the Protein Structure:==== | ====Analyzing the Protein Structure:==== | ||
| − | Open your protein.pdb in Chimera | + | Open your protein.pdb in Chimera. |
| + | |||
| + | '''Assess the protein structure:''' | ||
1. Are there any missing loops? | 1. Are there any missing loops? | ||
| Line 14: | Line 16: | ||
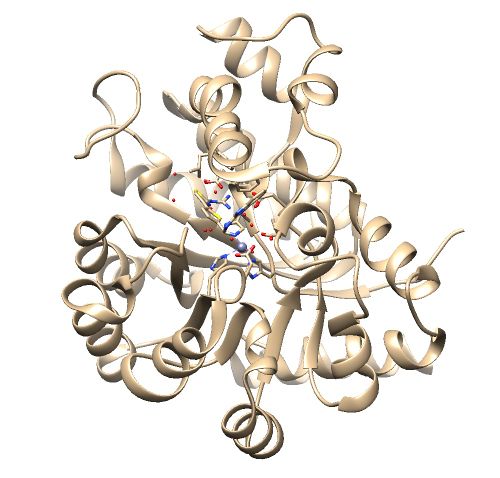
The .pdb for 1ndv from the RCSB Protein Data Bank is shown below. | The .pdb for 1ndv from the RCSB Protein Data Bank is shown below. | ||
| − | [[File:1ndv_raw_pdb.jpg|center | + | [[File:1ndv_raw_pdb.jpg|center|500px]] |
| + | |||
| + | '''Assessment:''' | ||
| + | |||
| + | 1. No missing loops. | ||
| + | |||
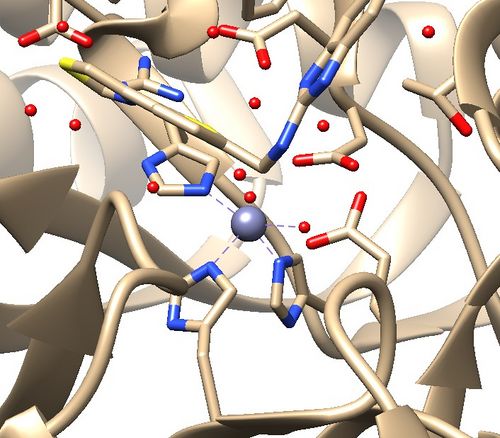
| + | 2. Zinc coordination with the protein and a water molecule. This zinc atom must be kept in the protein prep. | ||
| − | |||
| − | [[File:1ndv_coordination.jpg|center | + | [[File:1ndv_coordination.jpg|center|500px]] |
=== Ligand === | === Ligand === | ||
Revision as of 16:47, 6 March 2024
Contents
Introduction
Required Software
File Set-up
Preparing Structure Files
Protein
Analyzing the Protein Structure:
Open your protein.pdb in Chimera.
Assess the protein structure:
1. Are there any missing loops?
2. Are there any metal coordination atoms forming key interactions with the protein?
The .pdb for 1ndv from the RCSB Protein Data Bank is shown below.
Assessment:
1. No missing loops.
2. Zinc coordination with the protein and a water molecule. This zinc atom must be kept in the protein prep.