Difference between revisions of "2024 DOCK tutorial 1 with PDBID 2ITO"
Stonybrook (talk | contribs) (→Fixed Anchor Docking) |
Stonybrook (talk | contribs) (→Fixed Anchor Docking) |
||
| Line 166: | Line 166: | ||
[[File:2tio_fixed_anchor_out.png|thumb|center]] | [[File:2tio_fixed_anchor_out.png|thumb|center]] | ||
| + | |||
| + | The steps for the next docking mode should also be similar. | ||
===Flexible Docking=== | ===Flexible Docking=== | ||
Revision as of 16:25, 11 March 2024
Contents
- 1 Introduction
- 2 Preparation of the ligand and protein
- 3 Creating the Protein Binding Site Surface
- 4 Box and Grid Generation
- 5 Energy Minimization
- 6 DOCK
- 7 Virtual Screening of a Ligand Library
- 8 Cartesian Minimization of Virtually Screened Small Molecules
- 9 Rescoring and Ranking Virtually Screened Molecules
Introduction
For Numbered list do this:
- S
- D
- D
- P
- P
- F
These website tutorials are useful if you are unfamiliar with the following tools:
This tutorial is for PDBID 2ITO. Replace any reference to 2ITO with the protein of your choice.
Learning Objectives
These are for bullets:
- U
- G
Setting Up Your Environment
Sample picture from last year:
Downloading a protein from the PDB database
Preparation of the ligand and protein
Evaluating the Structure
Preparing the Protein file
Preparing the Ligand File
Final Steps
Creating the Protein Binding Site Surface
Creating the Required Surface (DMS) File
Generating Spheres for the Binding Site
Binding Site Spheres
Box and Grid Generation
Generating the Box
T
Generating the Grid
N
Energy Minimization
At
Ligand Minimization
Footprint Analysis
DOCK
Rigid Docking
W
Fixed Anchor Docking
In this session, we will be showing the tutorial for Fixed Anchor Docking. For this mode of docking, the algorithm would first identify the largest fragment extracted from original ligand input. Noted that, the fragments are segmented based on the rotatable bonds of the ligands. Starting from this fragment pose, the algorithm will use it as an anchor to continuously add segments until the whole original ligand pose has been fully re-grown. Noted that when each fragment is added, it would be oriented for a pose that is energetically minimal. Comparing this method to the previous rigid docking mode, fixed anchor docking offer flexibility to the side chain while still being able to keep the general location of the ligand’s binding site as it fixes the anchor position.
Please navigate to the directory designated for this docking mode: 007.fixed_anchor_docking. Here, we can start create the input parameters, file name – fixed.in:
vi fixed.in
The following is the input parameter for fixed anchor docking. While you can copy and paste this into the body of your script, please make sure to change the input files directory (ligand and protein structure folder) to your personal directory. Also it should be a good practice to make sure the directories for the dock parameters should also be up-to-date.
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/2tio.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.energy_min/2tio.lig.min_scored.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 2tio_fixed_output write_orientations no num_scored_conformers 1 rank_ligands no
Now, you are ready to run dock6 implementing the above input parameters:
dock6 -i fixed.in -o fixed.out
Dock6 should be running for a couple of minutes. If the program finished right away, it would be recommended that you should check the output file to see if there are any reported errors. Successful run should give you two additional files as followed:
- fixed.out
- 2tio_fixed_output_scored.mol2
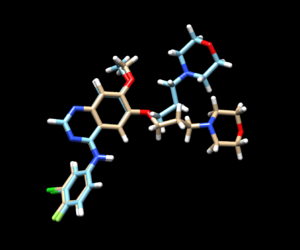
The mol2 file containing the docked ligand could be viewed in ChimeraX under ViewDock. To compare with the reference pose, this mol2 file (blue) has been overlaid on top of the original ligand pose (gold). For this result, it can be seen that although the docking score for the docked ligand is comparable with other docking modes, the RMSD computed for the docked result was larger than 2A, which has been reflected by the pose where the sidechain to the right is shifted further away than in original structure.
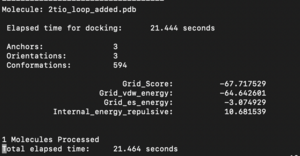
The fixed.out file summary the overall docking score of the run:
The steps for the next docking mode should also be similar.