Difference between revisions of "2018 Denovo design tutorial 2 with PDB 1C87"
(→Assembling your fragment library) |
|||
| Line 42: | Line 42: | ||
Once fraglib output files are created, make''denovo.in'' input file and run it interactively. | Once fraglib output files are created, make''denovo.in'' input file and run it interactively. | ||
| − | conformer_search_type | + | conformer_search_type denovo |
dn_fraglib_scaffold_file fraglib_scaffold.mol2 | dn_fraglib_scaffold_file fraglib_scaffold.mol2 | ||
dn_fraglib_linker_file fraglib_linker.mol2 | dn_fraglib_linker_file fraglib_linker.mol2 | ||
| Line 221: | Line 221: | ||
Now we will need an input file like before, but we will take a closer look at some of the parameters. In particular, we want to make sure we are using an appropriate amount of starting fragments (not too many or it will be a combinatorial explosion). | Now we will need an input file like before, but we will take a closer look at some of the parameters. In particular, we want to make sure we are using an appropriate amount of starting fragments (not too many or it will be a combinatorial explosion). | ||
| − | conformer_search_type | + | conformer_search_type denovo |
dn_fraglib_scaffold_file /PATH/fraglib_scaffold.mol2 | dn_fraglib_scaffold_file /PATH/fraglib_scaffold.mol2 | ||
dn_fraglib_linker_file /PATH/fraglib_linker.mol2 | dn_fraglib_linker_file /PATH/fraglib_linker.mol2 | ||
Revision as of 15:36, 6 September 2018
Contents
De novo design with PDB 1C87 focused fragment library
Preparing The Files
First of all, we need to create a directory for the fragment library.
mkdir fraglib
Move to fraglib directory and create an input file, fraglib.in
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix fraglib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy no ligand_atom_file /gpfs/projects/AMS536/2018/seung/001.files/lig_withH_charge_1c87.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix fraglib write_orientations no num_scored_conformers 1 rank_ligands no
fraglib_linker.mol2, fraglib_rigid.mol2, fraglib_scaffold.mol2, fraglib_scored.mol2, fraglib_sidechain.mol2 and fraglib_torenv.dat will be created. You can check how many linkers and side chains are in your system.
[seshin@login fraglib]$ grep -wc MOLECULE fraglib_linker.mol2 3 [seshin@login fraglib]$ grep -wc MOLECULE fraglib_sidechain.mol2 1
There are 3 linkers and 1 side chain. Once fraglib output files are created, makedenovo.in input file and run it interactively.
conformer_search_type denovo dn_fraglib_scaffold_file fraglib_scaffold.mol2 dn_fraglib_linker_file fraglib_linker.mol2 dn_fraglib_sidechain_file fraglib_sidechain.mol2 dn_user_specified_anchor yes dn_fraglib_anchor_file anchor1.mol2 dn_use_torenv_table yes dn_torenv_table fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 4 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump yes dn_write_orients no dn_write_growth_trees no dn_output_prefix 1c87.final use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../../002.spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../../004.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl
Create vi submit.sh in order to submit a job to create the output file.
#!/bin/bash #PBS -l walltime=48:00:00 #PBS -l nodes=1:ppn=24 #PBS -N 1c87_denovo #PBS -V #PBS -q long echo "Job started on" date cd $PBS_O_WORKDIR ../../../../zzz.programs/dock6_new/bin/dock6 -i denovo.in -o denovo.out echo "Job finished on" date
Denovo Rescore
Create a directory for denovo rescore.
mkdir denovo_rescore
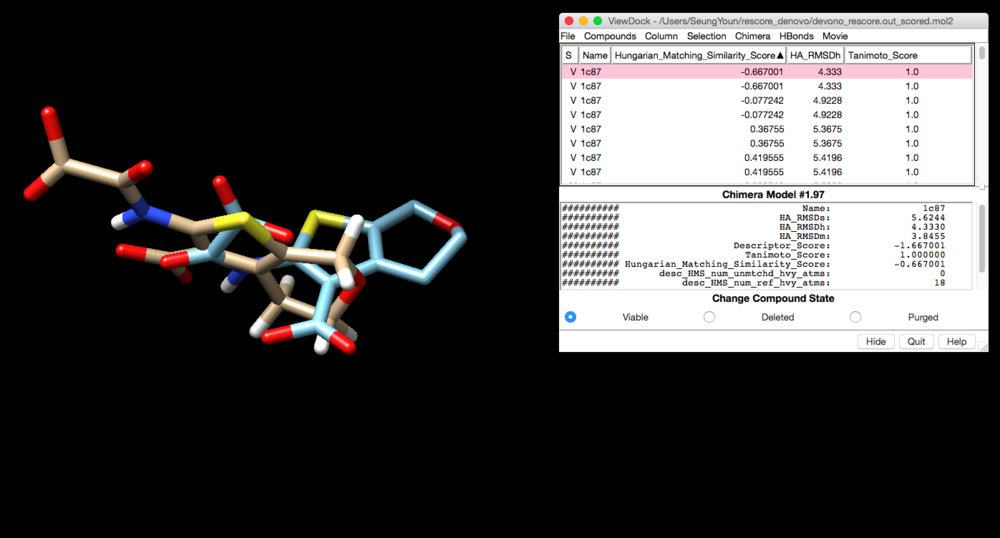
Make sure you have 1c87.final.denovo_build.mol2 file in your directory and lig_withH_charge_1c87.mol2 file will be your reference. (NOTE: Tantamoto is a chemical similarity. 1.0 means identical to the reference. )
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 1c87.final.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../../001.files/lig_withH_charge_1c87.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity no descriptor_use_pharmacophore_score no descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap no descriptor_fingerprint_ref_filename ../../001.files/lig_withH_charge_1c87.mol2 descriptor_hms_score_ref_filename ../../001.files/lig_withH_charge_1c87.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6/parameters/flex_drive.tbl ligand_outfile_prefix devono_rescore.out write_orientations no num_scored_conformers 1 rank_ligands no
devono_rescore.out_scored.mol2 will be created. Open Chimera and load lig_withH_charge_1c87.mol2 file and viewdock to open devono_rescore.out_scored.mol2
De novo design with PDB 1C87 generic fragment library
Assembling your fragment library
Actually, you won't be assembling anything. For this method we are using a generic set of fragments which we will use to grow our ligands. Like with the focused library, the traits of the cognate ligand will guide our designing algorithm so that we do not get anything too far-fetched.
Make a new directory for these calculations
mkdir denovo_growth_generic
Now we will need an input file like before, but we will take a closer look at some of the parameters. In particular, we want to make sure we are using an appropriate amount of starting fragments (not too many or it will be a combinatorial explosion).
conformer_search_type denovo dn_fraglib_scaffold_file /PATH/fraglib_scaffold.mol2 dn_fraglib_linker_file /PATH/fraglib_linker.mol2 dn_fraglib_sidechain_file /PATH/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table /PATH/fraglib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 300 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 4 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 2 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix out use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../surface-spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score yes descriptor_use_pharmacophore_score no descriptor_use_tanimoto no descriptor_use_hungarian no descriptor_use_volume_overlap no descriptor_multigrid_score_rep_rad_scale 1.0 descriptor_multigrid_score_vdw_scale 1.0 descriptor_multigrid_score_es_scale 1.0 descriptor_multigrid_score_number_of_grids 26 descriptor_multigrid_score_grid_prefix0 ../multi-grid/1c87.resid_023 descriptor_multigrid_score_grid_prefix1 ../multi-grid/1c87.resid_035 descriptor_multigrid_score_grid_prefix2 ../multi-grid/1c87.resid_044 descriptor_multigrid_score_grid_prefix3 ../multi-grid/1c87.resid_045 descriptor_multigrid_score_grid_prefix4 ../multi-grid/1c87.resid_046 descriptor_multigrid_score_grid_prefix5 ../multi-grid/1c87.resid_047 descriptor_multigrid_score_grid_prefix6 ../multi-grid/1c87.resid_048 descriptor_multigrid_score_grid_prefix7 ../multi-grid/1c87.resid_111 descriptor_multigrid_score_grid_prefix8 ../multi-grid/1c87.resid_114 descriptor_multigrid_score_grid_prefix9 ../multi-grid/1c87.resid_115 descriptor_multigrid_score_grid_prefix10 ../multi-grid/1c87.resid_119 descriptor_multigrid_score_grid_prefix11 ../multi-grid/1c87.resid_180 descriptor_multigrid_score_grid_prefix12 ../multi-grid/1c87.resid_181 descriptor_multigrid_score_grid_prefix13 ../multi-grid/1c87.resid_185 descriptor_multigrid_score_grid_prefix14 ../multi-grid/1c87.resid_214 descriptor_multigrid_score_grid_prefix15 ../multi-grid/1c87.resid_215 descriptor_multigrid_score_grid_prefix16 ../multi-grid/1c87.resid_216 descriptor_multigrid_score_grid_prefix17 ../multi-grid/1c87.resid_217 descriptor_multigrid_score_grid_prefix18 ../multi-grid/1c87.resid_218 descriptor_multigrid_score_grid_prefix19 ../multi-grid/1c87.resid_219 descriptor_multigrid_score_grid_prefix20 ../multi-grid/1c87.resid_220 descriptor_multigrid_score_grid_prefix21 ../multi-grid/1c87.resid_253 descriptor_multigrid_score_grid_prefix22 ../multi-grid/1c87.resid_256 descriptor_multigrid_score_grid_prefix23 ../multi-grid/1c87.resid_261 descriptor_multigrid_score_grid_prefix24 ../multi-grid/1c87.resid_264 descriptor_multigrid_score_grid_prefix25 ../multi-grid/1c87.resid_remaining descriptor_multigrid_score_fp_ref_mol no descriptor_multigrid_score_fp_ref_text yes descriptor_multigrid_score_footprint_text ../multi-grid/1c87.reference.txt descriptor_multigrid_score_foot_compare_type Euclidean descriptor_multigrid_score_normalize_foot no descriptor_multigrid_score_vdw_euc_scale 1.0 descriptor_multigrid_score_es_euc_scale 1.0 descriptor_weight_multigrid_score 1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file ../files/vdw_AMBER_parm99.defn flex_defn_file ../files/flex.defn flex_drive_file ../files/flex_drive.tbl