Difference between revisions of "2020 DOCK tutorial 4 with PDBID 6UZW"
Stonybrook (talk | contribs) (→Preparation of Receptor without Hydrogens) |
Stonybrook (talk | contribs) (→Preparation of ligand with Hydrogens) |
||
| Line 71: | Line 71: | ||
-Tools -> Surface Binding Analysis | -Tools -> Surface Binding Analysis | ||
Tools -> Structure Editing -> Add H (To add Hydrogen atoms) | Tools -> Structure Editing -> Add H (To add Hydrogen atoms) | ||
| − | Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force field available for standard residues. Here we used AMBER ff14SB) | + | Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force field |
| + | available for standard residues. Here we used AMBER ff14SB) | ||
Do not click or edit anything while going through this process. | Do not click or edit anything while going through this process. | ||
Save as a mol2 file. (6UZW_lig_withH.mol2) into 01_dockprep | Save as a mol2 file. (6UZW_lig_withH.mol2) into 01_dockprep | ||
| − | + | - If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor. | |
Tools -> Structure/Binding Analysis -> DockPrep | Tools -> Structure/Binding Analysis -> DockPrep | ||
Revision as of 13:43, 20 February 2020
This tutorial is created by AMS 536 Spring 2020 Group 4. Group 4 members include Steven Pak, Caitlyn Cardetti, MiaoMiao He and Chuanzhou (Joey) Zhu.
How to for selecting protein: Pick a protein with few substrates and rotational bonds?
The PDB code chosen is 6UZW which is for the crystal structure of GLUN1/GLUN2A ligand-binding domain in complex with glycine and UBP791.
Dock Prep Program used: Chimera
Contents
I. Introduction
DOCK
DOCK 6.9 is a molecular modeling program that identifies interactions between chemical compounds and the receptors. DOCK has many features that can help with drug discovery( ex: Virtual Screening and Denovo). This tutorial will show us how to use Virtual Screening.
6UZW
The tutorial will be based on the PDB file 6UZW downloaded from the PDB Database. 6UZW is the crystal structure for GLUN1/GLUN2A complexed with (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid and glycine.
Organization of Directories
We set up the files in our project space as such. It will be helpful to have these folders ready ahead of time.
00_files
01_dockprep
02_surface_spheres
03_gridbox
04_dock
05_footprint
06_virtual_screen
07_virtual_screen_mpi
08_cartesianmin
09_rescore
II. Preparation of the ligand and receptor (preparation before the usage of DOCK6.9
- Download 6UZW to local computer off of the PDB database - Save the .pdb file into your 00_files directory.
Checking the Structure and Preparing the Complex without Hydrogens
- Open Chimera, open the downloaded .pdb file, and check the structure for missing residues, gaps, heme groups, missing loops, and size. We recommend using proteins with no heme groups and proteins of a relatively smaller size. - Save the .pdb file (the ligand and the receptor) as a mol2 file (6UZW_complex_noH.mol2) - When preparing both the ligand the receptor, you can open this complex mol2 file and delete the component that you won't need (eg. for preparing the receptor, open the complex, delete the ligand, and save).
This complex (receptor and ligand) has been prepared without Hydrogens.
Preparation of Ligand without Hydrogens
- Open 6UZW_complex_noH.mol2 through Chimera again - Isolate the ligand by deleting the receptor. Select on the receptor by clicking on (Select --> Residue --> QM4 --> Select --> Inverted (selected models). The receptor should be surrounded by a green film. - Delete it (Actions -> Atoms/Bonds -> Delete). - The ligand should be by itself without any amino acids from the receptors and any water molecules. - Save the isolated ligand as a mol2 file (File -> Save mol2 -> 6UZW_lig_noH_mol2) into 01_dockprep
This ligand has been prepared without Hydrogens.
Preparation of Receptor without Hydrogens
- Open 6UZW_complex_noH.mol2 through Chimera again - Isolate the ligand by deleting the receptor. Select on the receptor by clicking on (Select --> Residue --> Standard Amino acids --> Select --> Inverted (selected models).
The ligand and water molecules should be surrounded by a green film.
- Delete it (Actions -> Atoms/Bonds -> Delete). - The the receptor should be by itself without the ligand - Save the isolated receptor as a mol2 file (File -> Save mol2 -> 6UZW_rec_noH_mol2) into 01_dockprep
This ligand has been prepared without Hydrogens.
Preparation of ligand with Hydrogens
-Open the 6UZW_lig_noH.mol2 -Tools -> Surface Binding Analysis
Tools -> Structure Editing -> Add H (To add Hydrogen atoms)
Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force field
available for standard residues. Here we used AMBER ff14SB)
Do not click or edit anything while going through this process.
Save as a mol2 file. (6UZW_lig_withH.mol2) into 01_dockprep
- If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor.
Tools -> Structure/Binding Analysis -> DockPrep
Preparation of receptor with Hydrogens
-Open the 6UZW_rec_noH.mol2 -Tools -> Surface Binding Analysis
Tools -> Structure Editing -> Add H (To add Hydrogen atoms)
Tools -> Structure Editing -> Add Charge (To add the charge use the latest AMBER force field available for standard residues. Here we used AMBER ff14SB)
Do not click or edit anything while going through this process.
Save as a mol2 file. (6UZW_rec_withH.mol2) into 01_dockprep
- If you follow the step below all the above stated steps will automatically appear one after the other to prepare the receptor.
Tools -> Structure/Binding Analysis -> DockPrep
III. Generating receptor surface and spheres
Preparation of DMS file
- Open 6UZW_rec_noH.mol2 using chimera. - Action -> Surface -> Show - Tools -> Structure Editing -> Write DMS - Save the 6UZW_rec_noH.dms into 02_surface_spheres folder
Reopen the file and make sure the surface was generated.
Transfer all the folders created so far to seawulf cluster to be used in DOCK using the Secure Copy command on unix from the local terminal that you have saved your files. NOT from the seawulf terminal
scp -r [FROM] [TO] scp -r PATH/folder/ PATH/seawulf_folder
Generating spheres
- Go to 02_surface_spheres folder - Create a new input file to create spheres by typing vim INSPH and type the following lines inside the file.
6UZW_rec_noH.dms R X 0.0 4.0 1.4 6UZW_rec.sph
The first line 6UZW_rec_noH.dms specifies the input file. R indicates that spheres generated will be outside of the receptor surface. X specifies all the points will be used. 0.0 is the distance in angstroms and it will avoid steric clashes. 4.0 is the maximum surface radius of the spheres and 1.4 is the minimum radius in angstroms.The last line 6UZW_spheres.sph creates a sphere file that contains clustered spheres.
Once the INSPH file is ready, type the following command to generate the spheres.
sphgen -i INSPH -o OUTSPH
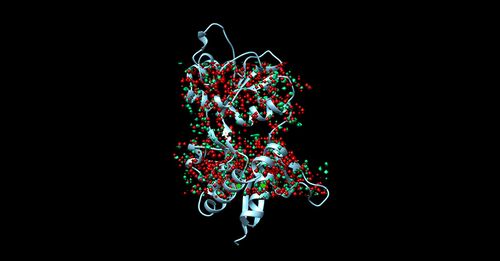
Once sphgen command is successful, 6UZW_rec.sph file will be created. Open it up using Chimera along with 6UZW_rec_noH.mol2 file. You should get a similar output like the image below.
Selecting Spheres
Here we will be selecting the spheres which define the binding pocket of the ligand because we are trying to direct the ligand towards that binding site rather than all over the receptor. To select the spheres type the following command.
sphere_selector 6UZW_rec.sph ../01_dockprep/6UZW_lig_withH.mol2 10.0
This command will select all of the spheres within 10.0 angstroms of the ligand and output them to selected_spheres.sph. Visualize the selected spheres using Chimera to make sure the correct spheres are selected. Notice that, spheres around the ligand-binding site are kept and all the other spheres are deleted in the image below.
Step 1. Separate protein and ligand
Step 2. Delete H2O molecules in Chimera to reduce computing time.
- To delete: Select > Residues > H O H - they should be highlighted Click Actions > Atoms/Bonds > delete H2O molecules should now be deleted.
Step 3. Add H's.
- PDB codes do not contain H's due to low electron density.
- To add H's Tools > Structure Editing > AddH
- H's MUST be added before you add charge or they will not be accounted for and will affect predictions for interactions.
Step 4. Add charge.
- Add charges Tools > Structure Editing > Add charge Typically use AM1-BCC
- You need to be careful about selecting charge because this will affect predictions for interactions (i.e. Coulomb's Law). Read paper corresponding to your PDB structure to determine conditions of purification and proper protonation. In the case of 6UZW our paper of reference is: https://doi.org/10.1038/s41467-020-14321-0
Pose Reproduction Flexible, Fixed-anchor, rigid
Generating the box and grid
For this part, we will be generating a box and grid for docking the ligand. A space-defined box will be searching efficiently, and a grid-based energy system is beneficial for a rapid and thoroughly process.
Step 1. Create a Box.
- Go to 03_gridbox folder - Create a new input file to create a box by typing vi showbox.in and type the following lines inside the file.
Y 8.0 ../02_surface_spheres/selected_spheres.sph 1 6UZW.box.pdb
Those are basically parameters/answers for questions specified in the program. The first line Y represents "Yes, we want to create a box around the sphere". 8.0 indicates that the box will have a length of 8 Angstroms, in all six directions. The third line specifies the sphere file should be used. 1 is the cluster number for creating the box. The last line 6UZW_box.pdb creates a file that contains the box.
Once the showbox.in file is ready, type the following command to create the box.
showbox < showbox.in
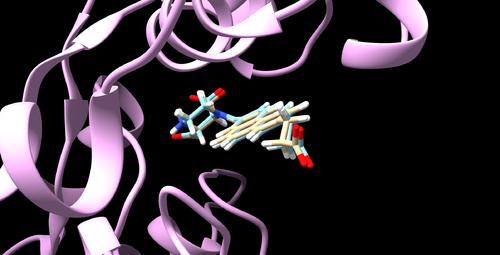
Once this command is successful, 6UZW.box.pdb file will be created. Open it up using Chimera and you should be able to visualize the structure like the image below.
Step 2. Generating the Grid
-In the same directory, create a new file by typing vi grid.in and type the following lines inside the file.
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file ../01_dockprep/6UZW_rec_prepped.mol2 box_file 6UZW.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Once the file is ready, type the following command to generate the grid. More information can be found in http://dock.compbio.ucsf.edu/DOCK_6/tutorials/grid_generation/generating_grid.html
grid -i grid.in -o grid.out
The grid calculation might takes a while to complete. But once this command is successful, four more files will be in 03_gridbox directory:
6uzw.box.pdb grid.in gridinfo.out grid.bmp grid.nrg showbox.in
Open it up using Chimera along with the receptor and make sure everything looks reasonable. If all is good, you should be able to visualize the structure like the image below.
IV.Energy Minimization and Footprint Analysis
Energy Minimization
Here before performing docking, we will do energy minimization for ligan to remove unfavorable clashes which influence how the rigid docking goes. Rigid docking need to have a better prepared ligand since the ligand will be docked as a whole, not like any other docking method which will take into account of several orientations and torsion angles while adding individual fragment.
Step 1. Create a input file.
-Go to the directory 04_dock -Create a new file by typing “touch min.in"
Step 2. Write into min.in file. For this part, we recommend you to write into the min.in file by answering them one by one instead of including content directly from previous. It’s a more safer way to keep the answers and questions correspond with each other, since the software update the question with time. Enter the command below:
dock6 -i min.in
The parameters you choose for those questions should be similar to the following:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../01_dockprep/6UZW_lig_prepped.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01_dockprep/6UZW_lig_prepped.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03_gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 6UZW_lig.min write_orientations no num_scored_conformers 1 rank_ligands no
If the process went smooth, a new file “ 6UZW_lig.min_scored.mol2 ” will be generated. You can visualize the structure along with receptor using Chimera and see how difference it is from the initial ligand.
Molecular Footprint
Molecular footprints is a commom way to present how a ligand interacts with the receptor in terms of electrostatic interactions and Van der Waals interactions. Here, we will generate a molecular footprint of the ligand interacts with the receptor before and after minimization.
Step 1. Create a new input file
-Go to directory 06_footprint -create an new file by typing “touch footprint.in"
Step 2. Write into min.in file.
Enter the command below:
dock6 -i footprint.in
Again, the file will be all set by answering a set of questions. The parameters you choose for those questions should be similar to the following:
conformer_search_type rigid use_internal_energy no ligand_atom_file ../04_dock/6UZW_lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ../01_dockprep/6UZW_lig_prepped.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ../01_dockprep/6UZW_rec_prepped.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 12 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 6UZW_footprint_min_cryst write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Once everything is successfully processed, several output files should be generated:
footprint.out 6UZW_footprint_min_cryst_footprint_scored.txt 6UZW_footprint_min_cryst_hbond_scored.txt 6UZW_footprint_min_cryst_scored.mol2
In order to visualize the result of molecular footprint. A Python command, which could be accessed in the previous DOCK tutorials, will be used to generate a PDF file. The PDF file will list the first 15 amino acid with a large deviations in energy.