Difference between revisions of "2022 DOCK tutorial 3 with PDBID 1X70"
BrockBoysan (talk | contribs) (→System Preparation) |
BrockBoysan (talk | contribs) (→System Preparation) |
||
| Line 8: | Line 8: | ||
'''Fetching 1X70''' | '''Fetching 1X70''' | ||
| − | Open Chimera: | + | Open Chimera and do the following to grab the protein: |
| − | File > Fetch By | + | File > Fetch By ID > 1X70 |
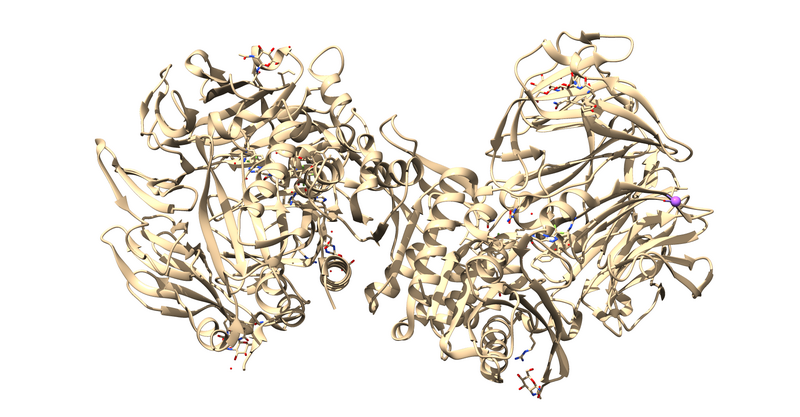
[[File:1X70_fetched.png|thumb|center|800px|Biological assembly 1 of 1X70.]] | [[File:1X70_fetched.png|thumb|center|800px|Biological assembly 1 of 1X70.]] | ||
| + | |||
| + | The first thing to notice is that this is a dimer and the ligand, 715, is not bound at the dimer interface. Thus, one of the monomers is entirely redundant and should be deleted. | ||
| + | |||
| + | Select > Chain > A | ||
| + | |||
| + | Actions > Atoms > Delete | ||
| + | |||
| + | Next it is important to remove cofactors, ions, and water molecules not involved in the binding interactions. This can be checked by reading through the paper associated with the PDB. | ||
| + | |||
| + | Select > Residue > NAG > Actions > Atoms > Delete | ||
| + | |||
| + | Select > Residue > NDG > Actions > Atoms > Delete | ||
| + | |||
| + | Select > Residue > HOH > Actions > Atoms > Delete | ||
| + | |||
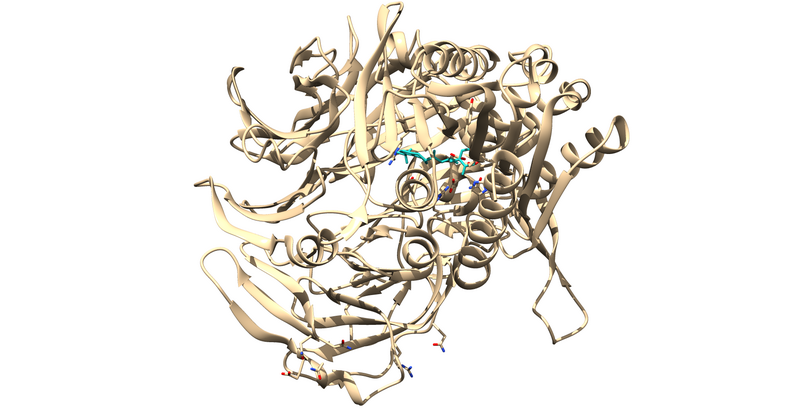
| + | This will leave us with just the ligand and receptor. | ||
| + | |||
| + | |||
| + | [[File:1X70_715_complexed.png|thumb|center|800px|1 monomer of 1X70 with all waters and non standard residues deleted except for the ligand 715, which is colored light blue for clarity.]] | ||
==Surface Generation & Spheres== | ==Surface Generation & Spheres== | ||
Revision as of 19:11, 28 February 2022
Contents
Introduction
DOCK
System Preparation
Fetching 1X70
Open Chimera and do the following to grab the protein:
File > Fetch By ID > 1X70
The first thing to notice is that this is a dimer and the ligand, 715, is not bound at the dimer interface. Thus, one of the monomers is entirely redundant and should be deleted.
Select > Chain > A
Actions > Atoms > Delete
Next it is important to remove cofactors, ions, and water molecules not involved in the binding interactions. This can be checked by reading through the paper associated with the PDB.
Select > Residue > NAG > Actions > Atoms > Delete
Select > Residue > NDG > Actions > Atoms > Delete
Select > Residue > HOH > Actions > Atoms > Delete
This will leave us with just the ligand and receptor.
Surface Generation & Spheres
This section details the generation of sphere files which will be used to describe where you are trying to DOCK to on your protein.
Surface Generation In Chimera: Load 1X70 w/o Hydrogens > actions > show > surface
Then you will write a DMS (Molecular Surface) File With the surface generated in Chimera: Tools > Structure Editing > Write DMS
Now you should have a DMS file for the next step.
Sphere Generation To generate spheres make the following input file: "INSPH" -
1X70_dms.dms #Molecular Surface File R #Whether to generate spheres outside of surface (R) or inside (L) X #Surface points from the DMS file to use in sphere generation 0 #Minimum radius between spheres 4.0 #Maximum radius of sphere 1.4 #Minimum radius of sphere 1X70_wo_H.sph #Output sphere file
For more information on sphere generation see: https://dock.compbio.ucsf.edu/DOCK_6/tutorials/sphere_generation/generating_spheres.htm
The .sph file should give you something similar to the following image if you load it up over your protein in Chimera:
Sphere Selection
Making The Infamous Grid
Making the grid
Energy Minimization for the ligand
Docking & Virtual Screening
Rigid Docking
Fixed Anchor Docking
Flexible Docking
Placeholder
[[File:|thumb|center|800px|image placeholder]]