Difference between revisions of "2023 DOCK tutorial 1 with PDBID 4S0V"
Stonybrook (talk | contribs) (→Creating the Required Surface (DMS) File) |
Stonybrook (talk | contribs) (→Generating Spheres for the Binding Site) |
||
| Line 148: | Line 148: | ||
===Generating Spheres for the Binding Site=== | ===Generating Spheres for the Binding Site=== | ||
| + | Now that we have a .dms file, we need to find the binding site of the protein using a DOCK program called sphgen. This program is run on the command line on Seawulf. In order for it to run properly you need to scp the .dms file to the 002.surface_spheres directory using the command: | ||
| + | scp filename.dms username@login.seawulf.stonybrook.edu:'/gpfs/projects/AMS536/2023/students/YOURGROUPNUMBER/002.surface_spheres' | ||
| + | |||
| + | fa | ||
===Selecting the Appropriate Spheres=== | ===Selecting the Appropriate Spheres=== | ||
Revision as of 20:36, 22 February 2023
Contents
Introduction
This tutorial will walk you through the steps necessary for using the DOCK software package. Many drugs are small molecular compounds that attach, or bind, to a protein in our bodies to change how that protein functions. By changing the function of a protein we can treat disease and help people manage symptoms of disorders. Traditionally drug discovery was done through a type of "trial and error" process called High Throughput Screening. Scientists would chemically make, or buy, thousands of small compounds and expose them to cells. They would then observe how the cells responded, either favorably/unfavorably/no effect. This method is time consuming and expensive. It would be better if the scientific community could "virtually screen" these molecules using a computer before creating/buying them - thereby focusing the cost and effort on those which showed the most promising computational results. The DOCK software brings this drug discovery process into the 21st century and uses computers to bind these small molecular compounds to a protein and evaluate the results. DOCK uses algorithms to bring together the small molecule, known as the ligand, and the larger protein, and "DOCK" them together. Our tutorial will walk you through preparing a protein and ligand for docking using an example complex from the protein data bank (https://www.rcsb.org/), complex # 4S0V.
The following steps will be followed:
- Setting up your environmnet
- Downloading a protein from the PDB database
- Determining if there are any missing loops and if they need to be modeled
- Preparing the ligand
- Preparing the protein
Learning Objectives
- Understand why DOCK was created and its current role in drug design
- Gain the ability perform virtual screening of small molecular compounds to a protein from the Protein Data Base (https://www.rcsb.org/)
Setting Up Your Environment
Before we get started working with molecules it's a good idea to set up your directory structure on Seawulf. To keep everything organized you need to create the following folder structure on Seawulf:
Downloading a protein from the PDB database
To begin we need to download protein complex #4S0V from the PDB. The right side of the top banner has a search bar:
.Simply type 4S0V into the search bar and the protein complex will be displayed.
On the right hand side, click on Download Files, then PDB Format.
That's it! The pdb file that we will be working with is now downloaded onto your local computer.
Preparation of the ligand and protein
The following steps will show you how to prepare the protein and ligand structures to be used with DOCK. All steps in this section will be done using Chimera. If you are unfamiliar with Chimaera, please see our tutorials here. These steps are very important - if your initial structure is not prepared properly, all downstream analysis can potentially be incorrect. This section will show you how to:
- Evaluate the structure to determine if there are any missing loops/water molecules to be removed/ions that need to be removed
- How to prepare the protein structure
- How to prepare the ligand structure
Evaluating the Structure
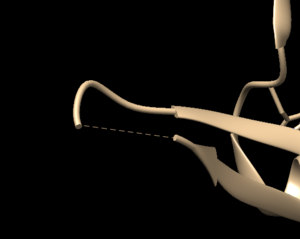
Open the previously downloaded .pdb file into Chimera. The first thing you want to look for are missing loops. A missing loop will be indicated by a dashed line in the structure:
The first decision you'll need to make is if these missing loops are important in your model or not. This decision is made by determining if the missing loop is close to the binding site. If it is far enough away, it probably won't affect the dynamics of the protein/ligand interaction and can be left alone. If the missing section is close to the binding site, you may want to fix it to more accurately model the binding site and the protein/ligand interaction.
To determine the distance between the missing loop and the binding site:
- Select an atom at the section of missing loop (ctrl)
- Select another atom near the binding site (ctrl + shift)
- Go to Tools → Structure Analysis → Distances
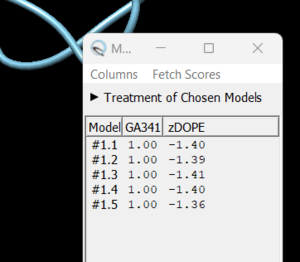
The distance between the two chosen atoms will appear if you click on 'create' in the dialogue box that pops up. If you determine that you want to re-create the missing sections, go to Tools → Surface Editing → Model/Refine Loops. A dialogue box will appear, choose 'non-terminal missing structure' and click 'Apply'. You can monitor the progress of Modeller in the lower left hand corner of the display. Once it has finished another dialogue box will appear showing you the five choices of models for the missing sections.
As you click on each of the results, the re-created missing section will show up. Decide which one you want to keep, highlight it and save the file by chooseing File → Save PDB. In the dialogue box, be sure to give this file a new name so as not to overwrite the original 4s0v.pdb file. In the 'Save models' section, choose the model number you chose above. Close the file and re-open Chimera. Now open the newly saved .pdb and you will no dashed lines and only the structure with the re-created loops.
You are now ready to move onto preparing the ligand and protein structures for docking.
Preparing the Protein file
The first step in preparing the protein is to get the protein structure alone in a .pdb file. To do this:
- Select an atom on the protein
- Press the up arrow until the entire protein is selected
- Go to Select → Invert (all models). This will change the selection from the protein to everything else in the structure
- Go to Actions → Atoms/Bonds → Delete
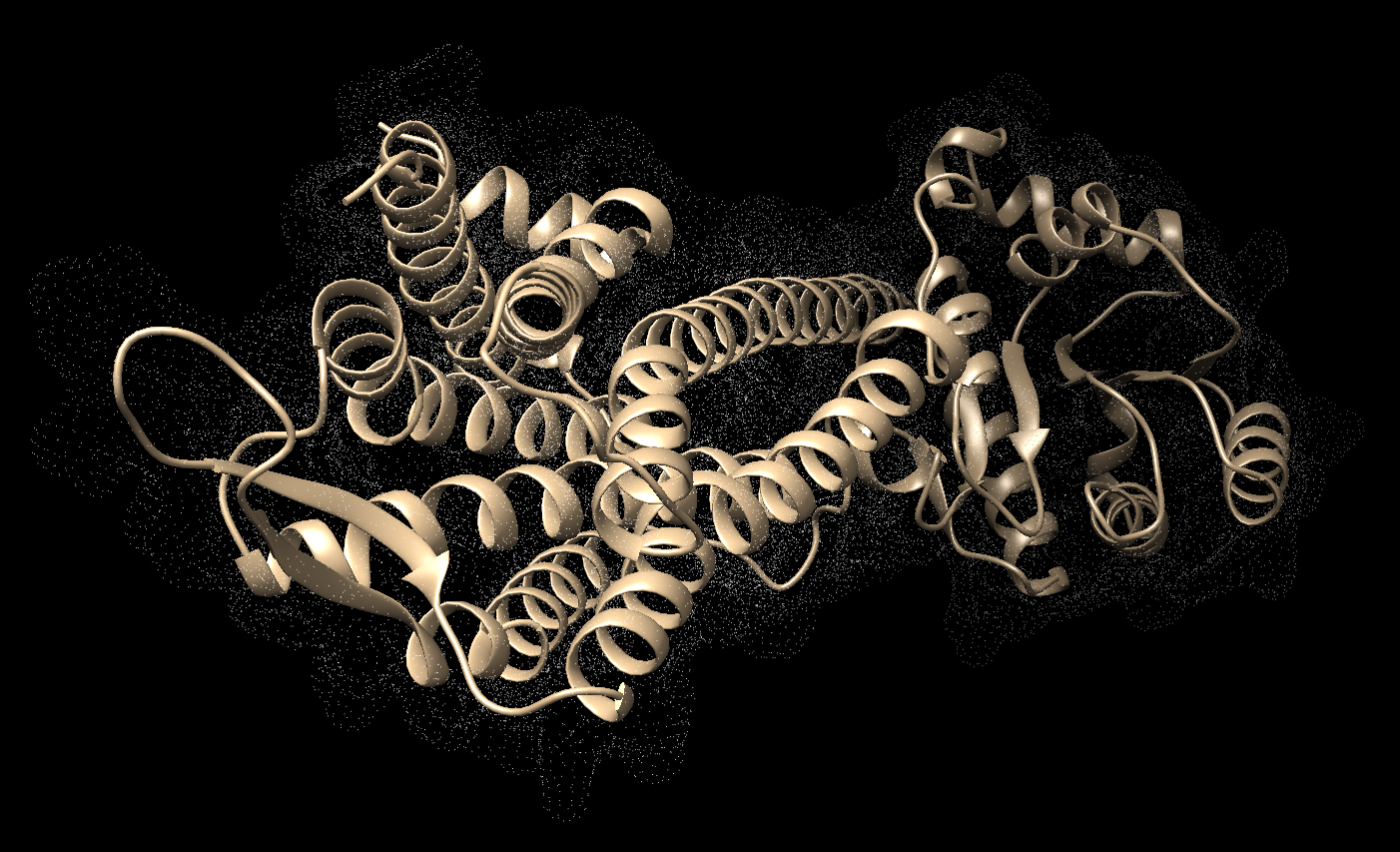
- Save the structure with a new file name (i.e. 4s0v_protein_only.pdb). Your pdb file will now look similar to this:
At this point we also need to generate a .mol2 file for the structure in this state. Go to File → Save Mol2 and give the file a descriptive name such as 4s0v_no_hydrogens_no_charges.mol2
There are now two more steps necessary for its preparation:
- Adding hydrogens
- Adding charge
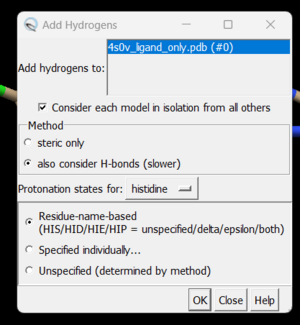
To add hydrogen click on: Tools → Structure Editing → AddH. This command will cause the following dialogue box to appear:
Click 'OK'. When the program is finished, the bottom left-hand side will say "Hydrogens added" but you won't see any change in the structure. It's good practice to make sure that the program worked properly by showing the atoms at a small area of the protein and ensuring hydrogens were added. To do with we're going to use the 'zone' command which is quite useful:
- Click on one atom anywhere on the protein
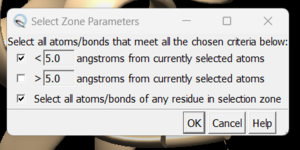
- Click on Select → Zone. This will cause the following dialogue box to appear:
make sure to click the same options as shown above and click on 'OK'. Go to Actions → Atoms/Bonds → show and the atoms in the atoms in the selected area will be shown. If the hydrogen atoms were successfully added you'll see structure similar to:
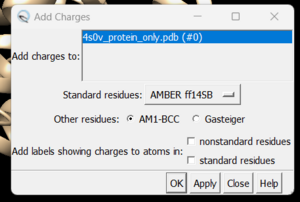
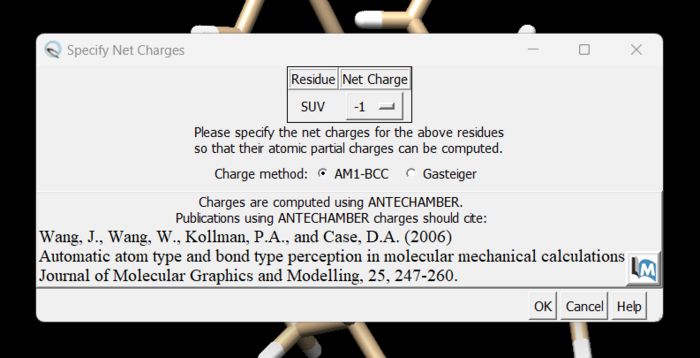
and you can see the white ends to the atoms which are the hydrogens. The final step is to add charges to the protein. Before you do this you should clear your selection by clicking on Select → Clear Selection. To add charges click on Tools → Structure Editing → Add Charge and the following dialogue box will show up:
Click on 'OK' and once the program is finished the bottom left hand corner will tell you what the total charge of the structure is. This number should be an integer. Your protein structure is now completely prepped and ready for docking. The final step is to save the file as a .mol2 file. Simply go to File → Save Mol2. Choose a descriptive file name such as 4s0v_protein_with_charges.mol2.
Preparing the Ligand File
The first step in preparing the ligand is the same as for the protein - we need to get the ligand structure alone in a .pdb file. To do this:
- Select an atom on the ligand
- Press the up arrow until the entire ligand is selected (you may have to press the up arrow many times)
- Go to Select → Invert (all models). This will change the selection from the ligand to everything else in the structure
- Go to Actions → Atom/Bonds → Delete
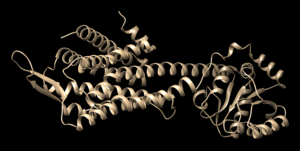
- Save the structure with a new file name (i.e. 4s0v_ligand_only.pdb). The image will look similar to this:
Before we start preparing the ligand for docking we need to save a .mol2 file for this structure with no hydrogens and no charges. Go to File → Save Mol2 and give the file a descriptive name such as 4s0v_no_hydrogens_no_charges.mol2
Once we have the ligand saved as its own file we follow the same two steps we did for the protein:
- Add hydrogens
- Add charges
These steps are a bit more complicated for the ligand because Chimera may have mistakes and we need to do our best to determine where hydrogens should be, what the overall charge of the ligand is, and what changes we need to make to the results presented by Chimera. Once the hydrogens are added to the ligand your file should be similar to:
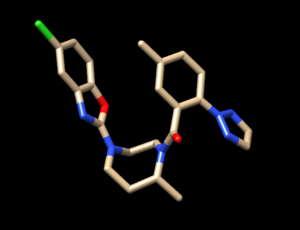
To determine if Chimera added hydrogens to the correct location we should look at the 2D structure of the ligand. This can be found on the pdb page where we downloaded the .pdb file.
clicking on the 2D image of the ligand brings up an enlarged image:
The first thing you'll want to look at are the carbons. Determine how many hydrogens should be attached to each and verify that's what Chimera added. Then move onto the oxygens and do the same thing. Next are the nitrogen atoms which are trickier. It's not always obvious if hydrogen atoms should be attached to the nitrogen atoms. A good way to determine this is to look at the original pdb file, with both the ligand and protein, with hydrogens added and determine if there are any interactions that don't seems right. To do this:
- Close your current session
- Open the original pdb that was downloaded
- Add hydrogens following the steps outlines above
- Look for interactions
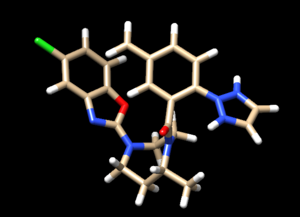
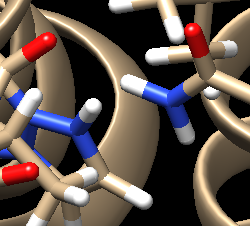
For the complex 4s0v, we see the following interaction that doesn't make sense:
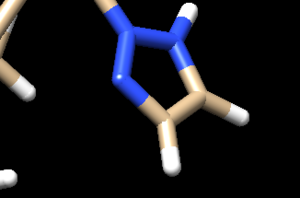
We see two hydrogens very close to each other. Hydrogens are positively charged so, in nature, if two were this close to each other they would repel each other, which isn't happening if the ligand is tightly bound to the protein. What we need to do is remove the hydrogen the Chimera put on the ligand. Hover your mouse over the hydrogen on the ligan, note it's number, and close the session. Open your ligand only file and select the hydrogen in question. Once the hydrogen is selected, go to Actions → Atom/Bonds → Delete which will delete the hydrogen we don't believe belongs there. Our ligand now looks like:
Looking at the ligand further we determined that the second hydrogen on the opposite nitrogen also didn't belong so that one was also removed. Once this was completed, the final step is to add charges to the ligand. We follow the same steps as for the protein except after the dialogue box appear and we click 'OK', a second box will appear:
and here a knowledge of organic chemistry will be needed. Chimera sees that we removed a hydrogen so expects the ligand charge to be -2 but the overall charge of the ligand should be 0. When the change is made in the dialogue box and we click 'OK' an error, the program runs successfully and we are done prepping the ligand for docking.
We just need to save the file as a .mol2 file. Go to File → Save Mol2 and give the file a name such as 4s0v_ligand_hydrogens_charges.mol2.
Creating the Protein Binding Site Surface
This section will walk you through the steps necessary to identify the binding site of the protein using a function within DOCK to place surface spheres along the protein.
Creating the Required Surface (DMS) File
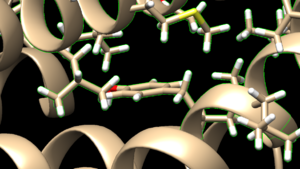
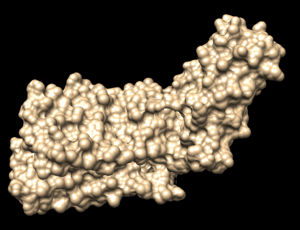
In Chimera, open the 4s0v_protein_only.pdb file. Go to Actions → Surface → show. This will display the van der Waals surface of your protein. Your image should be similar to:
Once this is done the .dms file needs to be generated by clicking on Tools → Surface Editing → Write DMS. A dialogue box will appear where you need to give the file a name, such as 4s0v_surface.dms. Once this file is saved, we need to make sure that nothing funny happened with DOCK during its generation. We do this by overlaying the .dms file over the protein_only file we previously saved. To do this, close your current session, open the 4s0v_protein_only.pdb and then open the 4s0v_surface.dms. If everything worked as it should you should see an image similar to:
As you can see, the small dots (which is the .dms file) is perfectly aligned over the protein structure. Now that we have verified that everything looks good, we can continue.
Generating Spheres for the Binding Site
Now that we have a .dms file, we need to find the binding site of the protein using a DOCK program called sphgen. This program is run on the command line on Seawulf. In order for it to run properly you need to scp the .dms file to the 002.surface_spheres directory using the command:
scp filename.dms username@login.seawulf.stonybrook.edu:'/gpfs/projects/AMS536/2023/students/YOURGROUPNUMBER/002.surface_spheres'
fa