Difference between revisions of "Test Set Construction SB2024 V1 DOCK6.10 A"
(→C. Receptor Preparation) |
|||
| Line 72: | Line 72: | ||
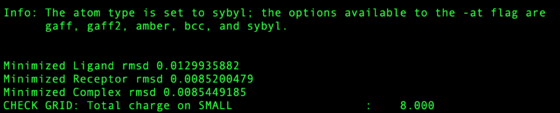
Check the output file for any large RMSD from amber minimization. Restraints in this step are set high so any RMSD > 0.5 Angstroms should be considered dubious. Any structure RMSD > 2.0 Angstroms should be rejected from the test set. | Check the output file for any large RMSD from amber minimization. Restraints in this step are set high so any RMSD > 0.5 Angstroms should be considered dubious. Any structure RMSD > 2.0 Angstroms should be rejected from the test set. | ||
| + | |||
| + | [[Image:Receptor rmsd ccorbo.png|thumb|center|560px|RMSD of Protein, Ligand and Complex for single system]] | ||
Due to an error in an Amber translation table P.3 atoms are converted to C.3 atoms when written to ${pdb_id}.rec.clean.mol2. At this point it is currently corrected manually, but is to be corrected in Amber23. | Due to an error in an Amber translation table P.3 atoms are converted to C.3 atoms when written to ${pdb_id}.rec.clean.mol2. At this point it is currently corrected manually, but is to be corrected in Amber23. | ||
Revision as of 16:24, 16 February 2024
!!!!!!Under Construction!!!!!!
The purpose of this tutorial is to develop a uniform method for adding systems to the Rizzo Lab test set, and rebuilding the set from initial files. This is the protocol to be used by all lab members. If the protocol is changed, a new tutorial should be made, preserving this tutorial.
Contents
- 1 I.Introduction
- 2 II.Scripts for System Preparation
- 3 A. Preliminary File Preparation
- 4 B. Ligand Preparation
- 5 C. Receptor Preparation
- 6 D. Sphere Generation
- 7 E. Single Grid Generation
- 8 F. Multi Grid Generation
- 9 G. Bookkeeping of Heavy Atoms
- 10 III.Test Cases for Preparation Integrity
- 11 IV.To Do
I.Introduction
A key development for this tutorial is bookkeeping scripts to manage the count of heavy atoms during processing and indicate unaccounted changes. Test cases are also included which should be visually inspected for indicated elements to be sure that processing occurred as expected.
II.Scripts for System Preparation
Each script should process the system in a fully automated manner. These scripts are located at (https://github.com/rizzolab/Testset_Protocols.git). There are key elements to check after each script before proceeding to the next. The output to be checked from each step is written to a folder "zzz.outfiles" with a unique file for each system, at each step. At the top of this file is the netid of the user who ran each process.
These scripts are designed to run in parallel on an HPC. Each step below has approximate timing indicated. If running 100 structures on a step with a timing of 10 minutes, at least 1000 cpu-minutes should be requested. There are averages and individual systems may take much less or more time.
A. Preliminary File Preparation
The receptor and ligand should be prepared from the initial PDB file as outlined in student provided VS tutorials (https://ringo.ams.stonybrook.edu/index.php/Tutorials). Briefly:
(1) The ligand should have hydrogens added with careful attention to protonation state and gasteiger charges added. The extracted ligand is saved as "${pdb_id}.lig.moe.mol2" .
(2) The receptor is saved in isolation. Generally any water molecules are deleted, except specialized experiments. Any atomic ions within 8 angstroms of the ligand are retained as part of the receptor. The protocols have prep and frcmod files integrated for Heme cofactors, so these should be retained as part of the receptor. Once the receptor, metal ions, and heme cofactor are extracted they are saved together as "${pdb_id}.rec.foramber.pdb"
(3) Any biologically relevant organic small molecule cofactor (such as NADH / NADPH) other than Heme should be treated as a ligand (1) and saved as "${pdb_id}.cof.moe.mol2".
(4) Files from (1) (2) and (3) should be moved to the folder zzz.master in the upper folder Testset_Protocols from the github repo.
For further clarification, review the student tutorials as indicated.
B. Ligand Preparation
Timing=10 minutes
This step will assign am1bcc charges based on the charge in the preliminary file ${pdb_id}.lig.moe.mol2 and be saved as ${pdb_id}.lig.am1bcc.mol2
In the script submit.run.001.sh, if using a list of PDB codes other than "zzz.lists/clean.systems.all", change this in the script first.
sbatch submit.run.001.sh
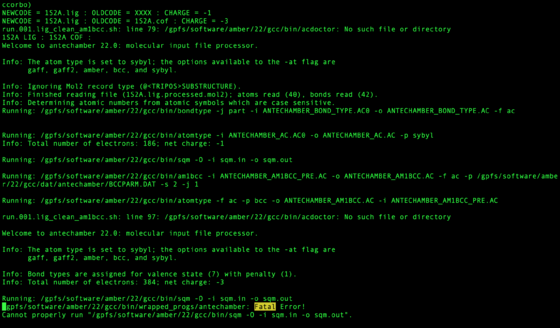
After the job has completed check to see if any of the ligands did not process successfully.
bash run.001b.check.sh
Any ligands or cofactors which did not pass will be listed in:
zyy.01.lig_missing.dat / zyy.01.cof_missing.dat
The output for each system is written to file with the netid of the user at the top:
vi zzz.outfiles/${pdb_id}.001.lig.out
These files should be checked for the phrase Fatal. This may mean the ligand failed or may have another issue. It does not mean with certainty the molecule is wrong, but should be inspected.
grep "Fatal" zzz.outfiles/*001.lig.out
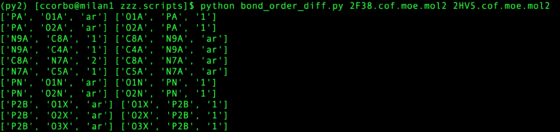
Some cofactors require unexpected bond orders to pass antechamber. If there is a correct version of the cofactor with the same res id such as "NAP" in zzz.master which passes antechamber, use this as a template.
python bond_order_diff.py ${pdb_id_1}.cof.moe.mol2 ${pdb_id_2}.cof.moe.mol2
This assumes atom names are equivalent for the mol2 (Hydrogen can be disregarded). Any bonds with different orders are printed, preserving the order input at command line (first argument prints on left, second argument prints on right). The script does not fix the bond orders for you, but helps quickly assigns how to match template.
Other common problems are with carboxylic acid and nitro functional groups. It is best to assign these functional groups "ar" bond types.
C. Receptor Preparation
Timing=1 minute
sbatch submit.run.002.sh
After completion check for missing systems:
bash run.002b.check.sh
Check the output file for any large RMSD from amber minimization. Restraints in this step are set high so any RMSD > 0.5 Angstroms should be considered dubious. Any structure RMSD > 2.0 Angstroms should be rejected from the test set.
Due to an error in an Amber translation table P.3 atoms are converted to C.3 atoms when written to ${pdb_id}.rec.clean.mol2. At this point it is currently corrected manually, but is to be corrected in Amber23.
D. Sphere Generation
E. Single Grid Generation
F. Multi Grid Generation
G. Bookkeeping of Heavy Atoms
III.Test Cases for Preparation Integrity
These are cases which that need to be visually inspected to ensure key operations during preparation were executed. This step is crucial after building the test set in batch mode.
| System | Element | Details |
|---|---|---|
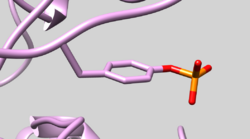
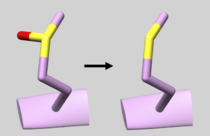
| 2GQG | Phosphotyrosine Residue | Check Residue 171 correctly has Y2P Phosphotyrosine (See Picture below) |
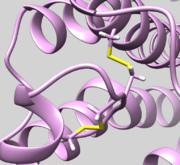
| 2Y03 | Disulfide Bond | Check bond Residue 82.SG and Residue 167.SG is actually bonded (See Picture below ) |
| 4WMZ | Heme | Check Heme is correctly incorporated, including Iron (Fe) |
| 1P44 | NADH Cofactor | Make sure cofactor is present in 1P44.rec.clean.mol2 and grid.out as residue "COF" |
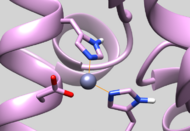
| 5UPG | Zinc Chelated Histidine Protonation | Residue should have HID protonation (See Picture below) |
| 3D94 | NonStandard Residue Defined Mutatation | Make sure Residue 146 successfully mutates to Residue "MET" from "MHO" |
| 1JWT | Long Bonds | Make sure TER is added before long bond |
-
-
IV.To Do
1. Integrate prep and frcmod files for cofactors available at http://amber.manchester.ac.uk/
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (2024)
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>