Difference between revisions of "2024 DOCK tutorial 3 with PDBID 1Y0X"
Stonybrook (talk | contribs) (→Fixed Anchor Docking) |
Stonybrook (talk | contribs) (→Flexible Docking) |
||
| Line 437: | Line 437: | ||
===Flexible Docking=== | ===Flexible Docking=== | ||
| + | The final docking method we're going to look at is called flexible docking. This is the most computationally expensive of the three methods, because now DOCK will sample all conformations of all rotatable bonds within the ligand attempting to minimize the energetics of the ligand. Once again we will be working on the command line in Seawulf, but now in your 008.flex_docking directory. | ||
| + | |||
| + | Create your input file: | ||
| + | vim flex.in | ||
| + | |||
| + | And type the following lines into your input file: | ||
| + | |||
| + | conformer_search_type flex | ||
| + | write_fragment_libraries no | ||
| + | user_specified_anchor no | ||
| + | limit_max_anchors no | ||
| + | min_anchor_size 5 | ||
| + | pruning_use_clustering yes | ||
| + | pruning_max_orients 1000 | ||
| + | pruning_clustering_cutoff 100 | ||
| + | pruning_conformer_score_cutoff 100.0 | ||
| + | pruning_conformer_score_scaling_factor 1.0 | ||
| + | use_clash_overlap no | ||
| + | write_growth_tree no | ||
| + | use_internal_energy yes | ||
| + | internal_energy_rep_exp 12 | ||
| + | internal_energy_cutoff 100.0 | ||
| + | ligand_atom_file ../004.energy_min/1y0x.lig.min_scored.mol2 | ||
| + | limit_max_ligands no | ||
| + | skip_molecule no | ||
| + | read_mol_solvation no | ||
| + | calculate_rmsd yes | ||
| + | use_rmsd_reference_mol yes | ||
| + | rmsd_reference_filename ../004.energy_min/1y0x.lig.min_scored.mol2 | ||
| + | use_database_filter no | ||
| + | orient_ligand yes | ||
| + | automated_matching yes | ||
| + | receptor_site_file ../002.surface_spheres/selected_spheres.sph | ||
| + | max_orientations 1000 | ||
| + | critical_points no | ||
| + | chemical_matching no | ||
| + | use_ligand_spheres no | ||
| + | bump_filter no | ||
| + | score_molecules yes | ||
| + | contact_score_primary no | ||
| + | grid_score_primary yes | ||
| + | grid_score_rep_rad_scale 1 | ||
| + | grid_score_vdw_scale 1 | ||
| + | grid_score_es_scale 1 | ||
| + | grid_score_grid_prefix ../003.gridbox/grid | ||
| + | minimize_ligand yes | ||
| + | minimize_anchor yes | ||
| + | minimize_flexible_growth yes | ||
| + | use_advanced_simplex_parameters no | ||
| + | minimize_flexible_growth_ramp yes | ||
| + | simplex_max_cycles 1 | ||
| + | simplex_score_converge 0.1 | ||
| + | simplex_initial_score_coverge 5 | ||
| + | simplex_cycle_converge 1.0 | ||
| + | simplex_trans_step 1.0 | ||
| + | simplex_rot_step 0.1 | ||
| + | simplex_tors_step 10.0 | ||
| + | simplex_anchor_max_iterations 500 | ||
| + | simplex_grow_max_iterations 250 | ||
| + | simplex_grow_tors_premin_iterations 0 | ||
| + | simplex_random_seed 0 | ||
| + | simplex_restraint_min no | ||
| + | atom_model all | ||
| + | vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn | ||
| + | flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn | ||
| + | flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl | ||
| + | ligand_outfile_prefix flex.out | ||
| + | write_orientations no | ||
| + | num_scored_conformers 10 | ||
| + | write_conformations yes | ||
| + | cluster_conformations yes | ||
| + | cluster_rmsd_threshold 2.0 | ||
| + | rank_ligands no | ||
| + | |||
| + | and run the file using the command: | ||
| + | dock6 -i flex.in -o flex.out | ||
| + | |||
| + | When the program has finished running you will see two new files in your directory: | ||
| + | *flex.out | ||
| + | *flex.out_scored.mol2 | ||
| + | |||
| + | |||
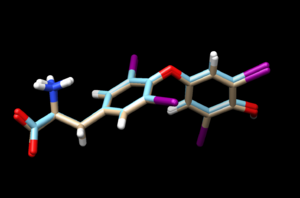
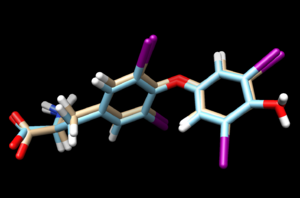
| + | again, scp the .mol2 file to your local computer and open it into a new session in Chimera. Overlaying this file with the energy minimized ligand we see: | ||
| + | [[File:1y0x flex.png|thumb|center]] | ||
= Virtual Screening of a Ligand Library = | = Virtual Screening of a Ligand Library = | ||
=Cartesian Minimization of Virtually Screened Small Molecules= | =Cartesian Minimization of Virtually Screened Small Molecules= | ||
=Rescoring and Ranking Virtually Screened Molecules= | =Rescoring and Ranking Virtually Screened Molecules= | ||
Revision as of 01:10, 18 March 2024
Contents
- 1 Introduction
- 2 Preparation of the ligand and protein
- 3 Creating the Protein Binding Site Surface
- 4 Box and Grid Generation
- 5 Energy Minimization
- 6 DOCK
- 7 Virtual Screening of a Ligand Library
- 8 Cartesian Minimization of Virtually Screened Small Molecules
- 9 Rescoring and Ranking Virtually Screened Molecules
Introduction
Proteins are mechano-chemical machines that governs many functions in cells and can caused serious diseases to human when malfunction. One active branch of protein research involves the design of small molecules that can bind to proteins at designated positions and inhibit/alter/modulate the function of the protein in a desirable manner. DOCK is a wonderful tool to be utilized to perform virtual screening (
- Setting up
- Preparing the protein and ligand
- Finding the binding site of the protein
- Generating the grid
- Ligand energy minimization
- Perform 3 common types of molecular docking
Learning Objectives
By following this tutorial, you will be able to successfully reproduce the docking result of the demonstrated case of 1Y0X and understand the fundamentals of virtual screening using DOCK6.10. Applying the same method illustrated in this tutorial for other protein system will likely yield meaningful results but there might be some slight fine tuning from case to case.
Preparation of the ligand and protein
- Evaluate the structure to determine if there are any missing loops
- Prepare the protein structure
- Prepare the ligand structure
Evaluating the Structure
- Select an atom at near the start of the missing section (hold the ctrl button while clicking it)
- Select another atom near the binding site (hold ctrl + shift while clicking the second atom)
- Go to Tools → Structure Analysis → Distances
Preparing the Protein file
- Select an atom on the protein
- Press the up arrow until the entire protein is selected
- Go to Select → Invert (all models). This will change the selection from the protein to everything else in the structure
- Go to Actions → Atoms/Bonds → Delete
- Save the structure with a new file name (i.e. 4s0v_protein_only.pdb). Your pdb file will now look similar to this:
- Adding hydrogens
- Adding charge
- Click on one atom anywhere on the protein
- Click on Select → Zone. This will cause the following dialogue box to appear:
Preparing the Ligand File
- Select an atom on the ligand
- Press the up arrow until the entire ligand is selected (you may have to press the up arrow many times)
- Go to Select → Invert (all models). This will change the selection from the ligand to everything else in the structure
- Go to Actions → Atom/Bonds → Delete
- Save the structure with a new file name (i.e. 4s0v_ligand_only.pdb). The image will look similar to this:
- Add hydrogens
- Add charges
Final Steps
Creating the Protein Binding Site Surface
Creating the Required Surface (DMS) File
Generating Spheres for the Binding Site
Binding Site Spheres
- scp selected_spheres.sph to your local computer
- Close any open sessions you have in Chimera
- In Chimera open selected_spheres.sph
- In the current session, open the original protein/ligand complex (4s0v.pdb)
- You should see the spheres located within the binding site of the protein, similar to:
- Hold down ctrl and click on a sphere
- Press the up arrow until all spheres are selected
- Actions → Atoms/Bonds → hide
- Verify the ligand is where the spheres were
Box and Grid Generation
The next step in the docking process is to generate energy interactions between the atoms of the protein and ligand. If this was done for the whole complex it would take too long to run to be useful. To get around this computationally expensive step, dock uses a box/grid method. We will define a box around the area of interest for the protein/ligand and DOCK will generate a grid within this box which will be used in the energy calculations.
Generating the Box
To generate the box we will be working again on the command line using a DOCK program called showbox. Start by logging into Seawulf and navigating to your 003.gridbox directory. We need to make a new file called showbox.in by typing:
vi showbox.in
This will create a new file, with a filename of showbox.in and open it in vi. The following commands need to be typed:
Y
8.0
../002.surface_spheres/selected_spheres.sph
1
1y0x.box.pdb
Remember to change the last line to be a filename with the number of protein you are working with. The second line of this code (8.0) is telling dock how many angstroms from the selected spheres to draw the box. Depending on your system you may need to modify this number.
To run this file, simply type:
showbox < showbox.in
If showbox was successful the file 1y0x.box.pdb will now be in your directory.
Generating the Grid
Now that we have our box defined we need to instruct DOCK to generate the grid within it. We do this using a DOCK program called grid:
vi grid.in
This command will generate and open a file named grid.in. The commands to be typed into this file are:
allow_non_integral_charges no compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4. bump_filter yes bump_overlap 0.75 receptor_file ../001.structure/protein_final.mol2 box_file 1y0x.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
The only change you need to make to the above commands is the receptor_file and box_file to reflect the files you previously generated.
Once this file is saved, run it:
grid -i grid.in -o 1y0xGridInfo.out
Be patient, this step might take a few minutes to run. You will know it's worked successfully if you see:
- grid.bmp
- grid.nrg
- 1y0xGridInfo.out
in your directory. With the box and grid successfully generated we are ready to move onto the energy minimization step.
Energy Minimization
At its core, DOCK is finding interactions between a protein and ligand by looking at energy interactions between atoms. In order for DOCK to give the most accurate results we need to ensure that the ligand is at its lowest energy state before docking it into the binding site of the protein.
Ligand Minimization
For this section we will be working in the 004.energy_min directory and will be using the dock6 command. Again we need to generate the input file that dock6 needs:
vim min.in
Once in vim the following lines need to be typed in:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.structure/ligand_final.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.structure/protein_final.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1y0x.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
And the file is run with the following command:
dock6 -i min.in -o min.out
After successful completion of the program two new files will be in your directory:
- min.out
- 1y0x.lig.min.mol2
scp the .mol2 file back to your local computer. To view the changes that dock made to the structure, and open the 1y0x.lig.min.mol2 file in Chimera and in the same session, open the ligand_final.pdb file. Remember the ligand_final.pdb is the file we saved after making the protonation changes to the ligand. You should see something similar to:
The 1y0x.ligand from PDB with hydrogen and charges is cyan and the newly generated and energy minimized 1y0x.lig.min.mol2 is tint
Footprint Analysis
At its core, DOCK can be thought of as a program which evaluates, minimizes, and uses the electrostatic and VDW interactions between the ligand and protein to determine how they bind to each other. A footprint analysis is a way to visualize these interactions and possibly be used to design new ligands with the same, or better, set of energetics. The steps in this section will be done on Seawulf in the 005.footprint directory. We will be using the dock6 command and again need to create an input file:
vi footprint.in
The following lines needed to be typed into the newly created file:
conformer_search_type rigid use_internal_energy no ligand_atom_file ../004.energy_min/1y0x.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary no multigrid_score_primary no dock3.5_score_primary no continuous_score_primary no footprint_similarity_score_primary yes fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ../001.structure/ligand_final.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ../001.structure/protein_final.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 9 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1y0x_footprint.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
to generate the footprint of the ligand/protein interaction type:
dock6 -i footprint.in
Again, you will know the program successfully ran if the following three files are now in your directory:
- 1y0x_footprint.out_footprint_scored.txt
- 1y0x_footprint.out_hbond_scored.txt
- 1y0x_footprint.out_scored.mol2
In order to view/analyze these results we need to use a python script that is located in the class directory. Copy it over to your 005.footprint directory with the following command:
cp /gpfs/projects/AMS536/zzz.programs/plot_footprint_single_magnitude.py .
To run this script, first we load the python using:
module load py/2.7.15
Then, type:
python plot_footprint_single_magnitude.py 1y0x_footprint.out_footprint_scored.txt 50
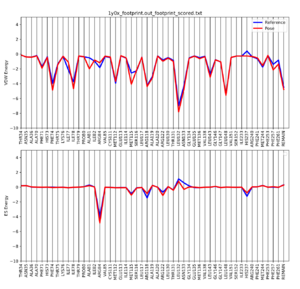
Once the script is completed you will see a .pdf file in your directory. scp this file back to your local computer. The file contains two plots that shows the energetic signature for the 50 most significant residues and will look similar to:
DOCK
We will be exploring three different options of the DOCK program:
- Rigid Docking
- Fixed Anchor Docking
- Flexible Docking
Each option has it's own pros and cons and your individual system should dictate which option you choose. Note: In the input file for each of these options has a line:
num_scored_conformers 1
Changing this number will change the number of conformations saved from the docking session. And for each options, to generate input file, the recommended way is to create a file first by typing:
touch example.in
then to make the file executable, type:
chmod+x example.in
then fill the input file using:
dock6 -i example.in
This will ask a few questions regarding the type of docking and other parameters and populate the input file accordingly. The input files for three different docking can be generated by answering those questions. In the next sections, the put files are pasted which was created from answering those questions.
Rigid Docking
When using rigid docking, the DOCK program does not sample different conformations of the ligand to try and find the most energetically stable. It simply treats the ligand as a rigid structure and tries to fit it into the binding site of the protein. For this step we will be using the energy minimized ligand file we generated above. This section will again be completed on Seawulf, please move to the 006.rigid_docking directory.
Again we need an input file that can be created using the command:
vi rigid.in
The following lines need to be typed into the file:
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/1y0x.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.energy_min/1y0x.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 10 write_conformations yes cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Remember to again change the necessary lines to point to your specific files. Once this is completed run the program by typing:
dock6 -i rigid.in -o rigid.out
Again, be patient as this may take a few minutes to complete. Once it's done running you will see two new files in your directory:
- rigid.out_scored.mol2
- rigid.out
scp the .mol2 file over to your local computer. Start a new session in Chimera and open the energy minimized ligand file from the previous step and the rigid docked ligand you just created (1y0x.lig.min_scored.mol2 and rigid.out_scored.mol2) to view the differences between the energy minimized ligand and DOCK's attempt to rigid dock that structure into the protein.
The 1y0x.lig.min_scored.mol2 is tint and the newly generated fixed.out_scored.mol2 is cyan.
Looking at this file we see that the two are very similar to each other.
Now we can look at the other file that dock generated, rigid.out. If we scroll to the bottom you will see grid scoring for this run:
Fixed Anchor Docking
The next DOCK option we will be looking at is called 'Fixed Anchor Docking'. For this option, DOCK cuts the ligand into fragments along its rotatable bonds. It then chooses the largest fragment as the 'anchor' and orients this fragment into the binding site of the protein. Once this is completed, the next fragments are added to the ligand, orientating them such that their energy is minimized but keeping them as rigid bodies. Comparing this to Rigid Docking, we see that more conformational sampling is being done but still within limits since each fragment is considered rigid.
We'll be working on the command line again, please move to the 007.fixed_anchor_docking directory and create an input file called fixed.in:
vim fixed.in
and type the following lines:
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/1y0x.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.energy_min/1y0x.lig.min_scored.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no minimize_flexible_growth_ramp no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/ vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 1y0x_fixed_output write_orientations no num_scored_conformers 1 rank_ligands no
Again, change the necessary lines to point to your specific files. To run this file type:
dock6 -i fixed.in -o fixed.out
Once it's done running you will see two new files in your directory:
- fixed.out
- 1y0x_fixed_output_scored.mol2
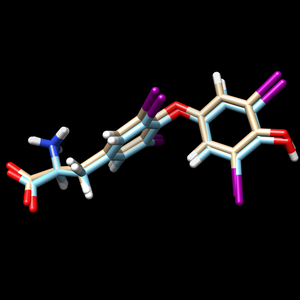
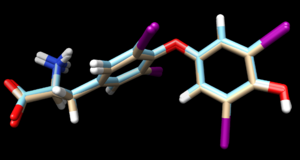
scp the .mol2 file over to your local computer. Start a new session in Chimera and open the energy minimized ligand file from the previous step and the fixed anchor docked ligand you just created (1y0x.lig.min_scored.mol2 and 1y0x_fixed_output_scored.mol2) to view the differences between the energy minimized ligand and DOCK's attempt to place that structure using its fixed anchor algorithm. Your image should be similar to:
The 1y0x.lig.min_scored.mol2 is cyan and the newly generated 1y0x_fixed_output_scored.mol2 is tint.
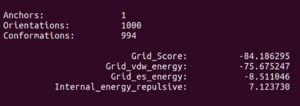
Now we can look at the other file that dock generated, fixed.out. If we scroll to the bottom you will see grid scoring for this run:
Anchors: 2
Orientations: 2
Conformations: 111
Grid_Score: -79.230011
Grid_vdw_energy: -73.074615
Grid_es_energy: -6.155398
Internal_energy_repulsive: 6.240775
Flexible Docking
The final docking method we're going to look at is called flexible docking. This is the most computationally expensive of the three methods, because now DOCK will sample all conformations of all rotatable bonds within the ligand attempting to minimize the energetics of the ligand. Once again we will be working on the command line in Seawulf, but now in your 008.flex_docking directory.
Create your input file:
vim flex.in
And type the following lines into your input file:
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../004.energy_min/1y0x.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../004.energy_min/1y0x.lig.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no minimize_flexible_growth_ramp yes simplex_max_cycles 1 simplex_score_converge 0.1 simplex_initial_score_coverge 5 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 250 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations no num_scored_conformers 10 write_conformations yes cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
and run the file using the command:
dock6 -i flex.in -o flex.out
When the program has finished running you will see two new files in your directory:
- flex.out
- flex.out_scored.mol2
again, scp the .mol2 file to your local computer and open it into a new session in Chimera. Overlaying this file with the energy minimized ligand we see: