Difference between revisions of "2024 Denovo tutorial 1 with PDBID 2ITO"
Stonybrook (talk | contribs) |
Stonybrook (talk | contribs) |
||
| Line 10: | Line 10: | ||
==Setting Up Your Environment== | ==Setting Up Your Environment== | ||
| − | + | We will add more directories to the ones we used from the traditional virtual screening. | |
| − | + | 009_deNovoRefinement | |
| + | 010a_fragLib | ||
| + | 010b_focusGrowth | ||
| + | 010c_focusReScore | ||
| + | 011_genericDeNovo | ||
=DeNovo Refinement= | =DeNovo Refinement= | ||
| − | + | DeNovo Refinement can be used to assess the effects upon ligand and protein interactions by chaning a portion of the molecule. We can experiment with an interesting piece of the ligand and delete the remaining portion. This deleted portion needs to be replaced with a dummy atom. Dock will then find which fragments can be placed in this position that will have the least energy. | |
==Setting up the dummy atom== | ==Setting up the dummy atom== | ||
Revision as of 20:41, 26 April 2024
Contents
Introduction
DeNovo Design aims to create a new ligand from scratch. It is incrementally designed bad added known structures of bioacitve compounds. The end result will hopefully produce a new ligand that can bind more tightly to a protein receptor than the current ligand can. There are three methods that will be introduced in this tutorial:
- Generic DeNovo Design
- Focused Fragment Design
- DeNovo Refinement
We will do denovo design with PDB: 2ITO.
Setting Up Your Environment
We will add more directories to the ones we used from the traditional virtual screening.
009_deNovoRefinement 010a_fragLib 010b_focusGrowth 010c_focusReScore 011_genericDeNovo
DeNovo Refinement
DeNovo Refinement can be used to assess the effects upon ligand and protein interactions by chaning a portion of the molecule. We can experiment with an interesting piece of the ligand and delete the remaining portion. This deleted portion needs to be replaced with a dummy atom. Dock will then find which fragments can be placed in this position that will have the least energy.
Setting up the dummy atom
For the ligand from #4s0v, we will be removing a terminal ring and looking at what DOCK suggests to replace it with. The steps to do this are:
- Open the ligand minimized mol2 file we generated in the previous tutorial into Chimera.
- Open the protein into the same session
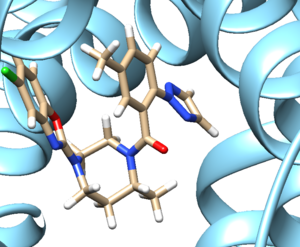
- Examine the binding site and choose a residue on the ligand that's pointing towards the inside of the binding site. For our protein this detailed section looks like:
We see an imidazole ring pointing towards the binding site so will choose to work with that. Select the protein and hide it from view:
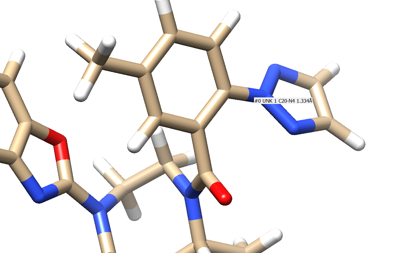
- Place your mouse over the atom connecting the ring to the rest of the ligand and note the atom and number. In this case it's N4.
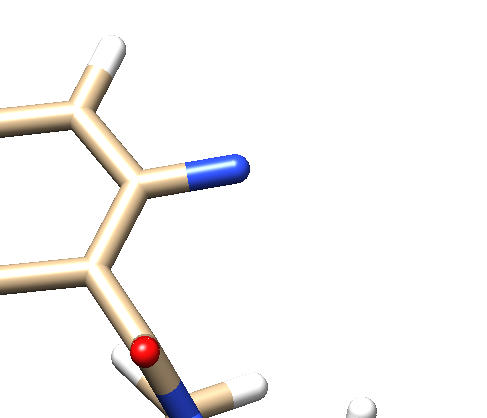
- Delete all the atoms from N4 to the end. Your ligand should now look something like:
- Save a .mol2 file of your ligand in this configuration. Make sure to give it a new filename such as 4s0v_denovoRefinement.mol2
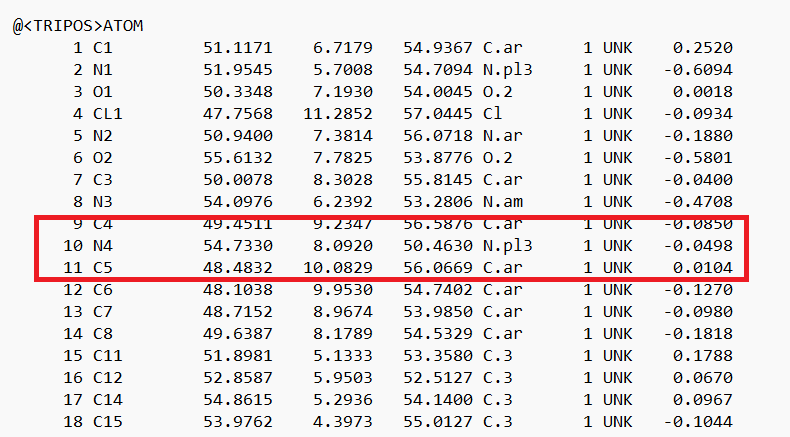
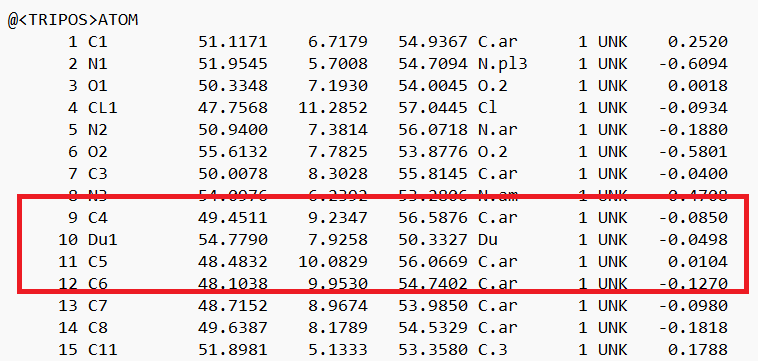
- Open the .mol2 file. If you're on a UNIX system, you can use vi; if you're on a PC, you can use textedit. Locate the atom that will now be changed to a dummy atom:
- Change the atom type to 'Du1' and the bond type to 'Du':
and save the file.
- Open a new session in Chimera and load the modified mol2 file. The "dummy" atom should now be purple:
- scp 4s0v_denovoRefinement.mol2 over to Seawulf and into the 012.denovoRefinement directory. From this point on we will be working on the command line.
Running DeNovo Refinement
Viewing New Molecules
Focused DeNovo Design
Fragment Library Generation
DeNovo Design
ReScoring Designed Molecules
Generic DeNovo Design
For generic denovo design, we are not limiting DOCK to build new molecules using fragments present in the original ligand. Instead, we will be taking fragments from a fragment library.
Create the input file:
vi 2ito_generic.in
The file should contain these lines:
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_torenv.dat dn_name_identifier 2ito_generic dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix 2ito_generic use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file /gpfs/projects/AMS536/2024/students/group_1_2ITO/002_surface_spheres/2ITO.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix /gpfs/projects/AMS536/2024/students/group_1_2ITO/003_gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 250 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
Create a slurm file to run this:
#!/bin/bash #SBATCH --job-name=2ito_generic #SBATCH --ntasks=28 #SBATCH --nodes=1 #SBATCH --time=48:00:00 #SBATCH -p long-28core dock6 -i 2ito_generic.in
After the program has run, you can use the following command to see how many ligands has been generated. The program has made 368 potential ligands.
grep MOLECULE 2ito_generic.denovo_build.mol2 | wc -l
The output file we are going to move to our local computer is 2ito_generic.denovo_build.mol2. We can see the ligands the program has created.
Go into Chimera and open the molecule on ViewDock. (Tools >> Surface/Binding Analysis >> ViewDock)
Use Column >> Show >> Grid_Score to find the ligand with the lowest energy.
The ligand with the lowest score has a score of -93.318.
We can compare this with the energy minimized ligand and see how it is positioned differently compared to the generated ligand. This original energy minimized ligand has a grid score of -71.423.
We can also observe how the designed ligand and the original ligand interact with the receptor.