Difference between revisions of "2024 AMBER tutorial 2 with PDBID 1NDV"
Stonybrook (talk | contribs) (→MM-GBSA) |
Stonybrook (talk | contribs) (→MM-GBSA) |
||
| Line 483: | Line 483: | ||
/ | / | ||
This is a complex calcualtion and should not be run on the head node. Create a submissions script called mmgbsa.sh and add the following lines: | This is a complex calcualtion and should not be run on the head node. Create a submissions script called mmgbsa.sh and add the following lines: | ||
| + | #! /bin/sh | ||
| + | #SBATCH --job-name=production_unrest | ||
| + | #SBATCH --ntasks-per-node=28 | ||
| + | #SBATCH --nodes=1 | ||
| + | #SBATCH --time=7-00:00:0 | ||
| + | #SBATCH -p extended-28core | ||
| + | cd $SLURM_SUBMIT_DIR | ||
| + | solv_parm="../001.tleap_build/1ndv.wet.complex.prmtop" | ||
| + | complex_parm="../001.tleap_build/1ndv.gas.complex.parm7" | ||
| + | receptor_parm="../001.tleap_build/1ndv.gas.receptor.parm7" | ||
| + | lig_parm="../001.tleap_build/1ndv.gas.ligand.parm7" | ||
| + | trajectory="../003.production/10_prod.trj" | ||
| + | MMPBSA.py -O -i mmgbsa.in \ | ||
| + | -o 1ndv_mmgbsa_results.dat \ | ||
| + | -eo 1ndv_mmgbsa_perframe.dat \ | ||
| + | -sp ${solv_parm} \ | ||
| + | -cp ${complex_parm} \ | ||
| + | -rp ${receptor_parm} \ | ||
| + | -lp ${lig_parm} \ | ||
| + | -y ${trajectory} | ||
Revision as of 22:20, 5 May 2024
Contents
Introduction
DOCK6 is a great tool that helps us understand ligand binding poses and relative binding energies. When DOCK6 is combined with another computational method known as molecular dynamics, we can calculate free energies of binding and probe how the ligand behaves in the binding pocket. In this tutorial we will walk through the use of AMBER16 to perform and analyze molecular dynamic simulations of PDBID 1NDV.
Getting Started
Before starting simulations it is important to create our directories so that we can keep our simulations organized. Create the following directories:
000.paramters 001.tleap_build 002.equilbration 003.production 004.analysis zzz.master
We will use the zzz.master directory to store our initial mol2 files and other files we may use that do not belong in another directory. Once you have your directories created you can begin to prepare they system for simulation.
Before Simulation
Structure
We will need mol2 and pdb files of the ligand and receptor separately for these simulations. If you followed the 1NDV DOCK6 VS tutorial you will already have these files and can move them to the zzz.master directory. If you did not follow that tutorial the steps will be listed below, but please refer to it if you require a more in depth tutorial. Please note that the receptor file should be without charges and hydrogens, while the ligand will need the charges and hydrogens.
We will start by generating our receptor file. Open the PDB of 1NDV in Chimera and follow these steps.
1) Select an atom on the receptor and use the entire up arrow until the entire protein is selected 2) Select -> Invert (all models) 3) Actions -> Atoms/Bonds -> delete
This should leave us with just the receptor atoms. Save this as a mol2 file and a pdb file as "1ndv_protein_noH_noCharge" DO NOT add charges or hydrogens.
We will now create the ligand file in a similar fashion.
1) Select an atom on the ligand and use the entire up arrow until the entire protein is selected 2) Select -> Invert (all models) 3) Actions -> Atoms/Bonds -> delete
Before we use this file we must add hydrogens and charges.
To add Hydrogens: Tools -> Structure Editing -> Add Hydrogens To add Charges: Tools -> Structure Editing -> Add Charges (for this ligand we will use an overall charge of 0)
Be sure to reference the PDB file to ensure that Chimera adds hydrogens in the correct places. Once charges and hydrogens are added you can save the mol2 and pdb files as "1ndv_ligand_addH_addCharge" Place both of these files in the zzz.master directory.
Force Field Parameters
Before we can use TLEap, we need to generate forcefield parameters for our ligand. We will use the antechamber program to do this. Move to the 000.paramters directory and use the following command to generate the forcefield parameter file.
antechamber -i ../zzz.master/1ndv_ligand_addH_addCharge.mol2 -fi mol2 -o 1ndv_ligand_antechamber.mol2 -fo mol2 -at gaff -c bcc -rn LIG
When running this you may encounter some warnings such as:
Warning: The number of bonds (3) for atom (ID: 1, Name: N1) does not match the connectivity (2) for atom type (N.ar) defined in CORR_NAME_TYPE.DAT.
Be sure to double check your ligand has the correct bond connectivity, and then these warnings may be safely ignored. You may also encounter some errors that prevent antechamber from running. It may require you to enter the mol2 file and edit the atom types to resolve them. Once antechamber has successfully run, run the following command to make our modified forcefield parameters:
parmchk2 -i 1ndv_ligand_antechamber.mol2 -f mol2 -o 1ndv_ligand.am1bcc.frcmod
TLEap
Now that we have our modified forcefield parameters we can use TLEap to solvate and prepare the system for simulation. Create a file called "tleap.build.in" and add the folowing commands:
#!/usr/bin/sh
###load protein force field
source leaprc.protein.ff14SB
###load GAFF force field (for our ligand)
source leaprc.gaff
###load TIP3P (water) force field
source leaprc.water.tip3p
###load ions frcmod for the tip3p model
loadamberparams frcmod.ionsjc_tip3p
###needed so we can use igb=8 model
set default PBradii mbondi3
###load protein pdb file
rec=loadpdb ../001.structure/1ndv_built.pdb
#THIS IS WHERE YOU WOULD DEFINE DISULFIDE BONDS
#NUMBERING SHOULD MATCH INPUT PDB FILE
#bond rec.Res#.SG rec.Res#.SG
###load ligand frcmod/mol2
loadamberparams ../002.parameters/1ndv_ligand.am1bcc.frcmod
lig=loadmol2 ../002.parameters/1ndv_ligand_antechamber.mol2
###create gase-phase complex
gascomplex= combine {rec lig}
###write gas-phase pdb
savepdb gascomplex 1ndv.gas.complex.pdb
###write gase-phase toplogy and coord files for MMGBSA calc
saveamberparm gascomplex 1ndv.complex.parm7 1ndv.gas.complex.rst7
saveamberparm rec 1ndv.gas.receptor.parm7 1ndv.gas.receptor.rst7
saveamberparm lig 1ndv.gas.ligand.parm7 1ndv.gas.ligand.rst7
###create solvated complex (albeit redundant)
solvcomplex= combine {rec lig}
###solvate the system
solvateoct solvcomplex TIP3PBOX 12.0
###Neutralize system
addions solvcomplex Cl- 0
addions solvcomplex Na+ 0
#write solvated pdb file
savepdb solvcomplex 1ndv.wet.complex.pdb
###check the system
charge solvcomplex
check solvcomplex
###write solvated toplogy and coordinate file
saveamberparm solvcomplex 1ndv.wet.complex.prmtop 1ndv.wet.complex.rst7
quit
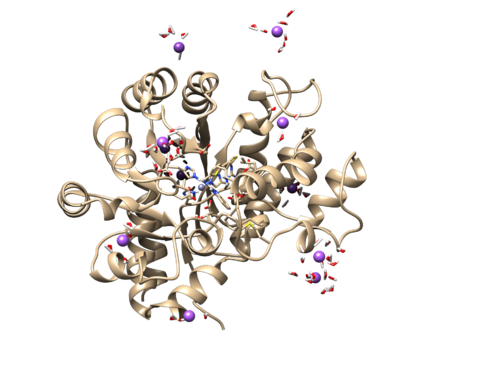
Be sure that all files are generated, especially the wet.complex files, and review the tleap_output.txt file to ensure that no errors were encountered. We will also load the wet.complx files into Chimera to make sure TLEaP ran properly. Download the 1ndv.wet.complex.prmtop and 1ndv.wet.complex.rst7. Now in Chiemra:
Tools → MD/Ensemble Analysis → MD Movie
We will use the rst7 file as our trajectory for this step. It should load an image similar to this.
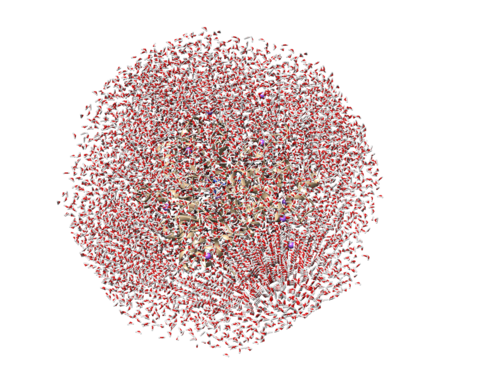
Now lets make sure that the rest of the waters were added properly and that our complex is in the solvent box. To do this:
1) Select → Chain → water 2) Actions → Atoms/Bond → Show
This should show all the waters added by TLEaP and should look similar to this.
Minimization and Equilibration
Before we run a production simulation, we must first minimize and equilibrate the system. We will do this with a 9-step procedure. This process will minimize the energy of the structure, slowly heat the system to the desired temperature, and then equilibrate the system at that temperature. First navigate the the 002.equiilibration directory. As we create the input files pay attention to the "restrainmask" variable. We want to restrain the protein and ligand atoms but not the hydrogens added by TLEaP so this will be set to ":1-350 & !@H=" for our system but be sure to change this is you are using a different receptor and ligand. This will change to just the protein atoms i.e. ":1-349 & !@H=" after step 7.
Create a file 01.min.mdin and add the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=5000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create a file 02.equil.mdin and add the following lines:
MD simualation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-235 & !@H=", ! atoms to be restrained restraint_wt=5.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create a file 03.min.mdin and add the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=2.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create a file 04.min.mdin and add the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create a file 05.min.mdin and add the following lines:
Minmize all the hydrogens &cntrl imin=1, ! Minimize the initial structure maxcyc=1000, ! Maximum number of cycles for minimization ntb=1, ! Constant volume ntp=0, ! No pressure scaling ntf=1, ! Complete force evaluation ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=0.05, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format /
Create a file 06.equil.mdin and add the following lines:
MD simualation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntb=2, ! Constant Pressure ntc=1, ! No SHAKE on bonds between hydrogens dt=0.001, ! Timestep (ps) ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=1.0, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create a file 07.equil.mdin and add the following lines:
MD simulation &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-350 & !@H=", ! atoms to be restrained restraint_wt=0.5, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create a file 08.equil.mdin and add the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-349@CA.C.N", ! atoms to be restrained, only the backbone restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
Create a file 09.equil.mdin and add the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=50000 ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=1, ! No SHAKE on for bonds with hydrogen dt=0.001, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=1, ! Complete force evaluation ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 1000, ! Write to trajectory file every ntwx steps ntpr= 1000, ! Print to mdout every ntpr steps ntwr= 1000, ! Write a restart file every ntwr steps cut= 8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-349@CA.C.N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on /
This should not be run on the head node, instead create a submission script called equil.sh and add the following lines:
#! /bin/sh
#SBATCH --job-name=sys_equilibration
#SBATCH --ntasks-per-node=40
#SBATCH --nodes=2
#SBATCH --time=8:00:00
#SBATCH -p long-40core
cd $SLURM_SUBMIT_DIR
echo "Started Equilibration on `date` "
do_parallel="mpirun pmemd.MPI"
prmtop="../001.tleap_build/2nnq.wet.complex.prmtop"
coords="../001.tleap_build/2nnq.wet.complex"
MDINPUTS=(01.min 02.equil 03.min 04.min 05.min 06.equil 07.equil 08.equil 09.equil)
for input in ${MDINPUTS[@]}; do
$do_parallel -O -i ${input}.mdin -o ${input}.mdout -p $prmtop -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r
${input}.rst7
coords=$input
done
echo "Finished Equilibration on `date` "
Finally run equilibrations using:
sbatch equil.sh
This simulations will take some time. Once all steps have completed, be sure to check the output.txt files and ensure they are reasonable.
Production
Now the system is ready for production. Production runs can be run with restrains on the protein, or without restraints on the protein. In this tutorial we will keep the restrains on the protein but this can be changed by removing the "restraintmask" and "restraint_wt" variables. Now navigate to the 003.production directory.
Create a file 10.prod.mdin and add the following lines:
MD simulations &cntrl imin=0, ! Perform MD nstlim=5000000, ! Number of MD steps ntx=5, ! Positions and velocities read formatted irest=1, ! Restart calculation ntc=2, ! SHAKE on for bonds with hydrogen dt=0.002, ! Timestep (ps) ntb=2, ! Constant Pressure ntp=1, ! Isotropic pressure scaling barostat=1 ! Berendsen taup=0.5 ! Pressure relaxtion time (ps) ntf=2, ! No force evaluation for bonds with hydrogen ntt=3, ! Langevin thermostat gamma_ln=2.0 ! Collision Frequency for thermostat ig=-1, ! Random seed for thermostat temp0=298.15 ! Simulation temperature (K) ntwx= 2500, ! Write to trajectory file every ntwx steps ntpr= 2500, ! Print to mdout every ntpr steps ntwr= 5000000, ! Write a restart file every ntwr steps cut=8.0, ! Nonbonded cutoff in Angstroms ntr=1, ! Turn on restraints restraintmask=":1-349@CA,C,N", ! atoms to be restrained restraint_wt=0.1, ! force constant for restraint ntxo=1, ! Write coordinate file in ASCII format ioutfm=0, ! Write trajectory file in ASCII format iwrap=1, ! iwrap is turned on $end
This simulation will also require a slurm script. This will take a very long time so make sure you select a queue that will allow it to finish.
Create a file prod.sh and add the following lines:
#! /bin/sh
#SBATCH --job-name=production_unrest
#SBATCH --ntasks-per-node=28
#SBATCH --nodes=1
#SBATCH --time=7-00:00:0
#SBATCH -p extended-28core
cd $SLURM_SUBMIT_DIR
echo "Started Production on `date` "
#do_parallel="mpirun pmemd.MPI"
do_parallel="/gpfs/software/intel/parallel-studio-xe/2018_3/compilers_and_libraries/linux/mpi/bin64/mpirun -np 80 /gpfs/software/amber/16/intel/cpu/bin/pmemd.MPI"
#do_cuda="pmemd.cuda"
prmtop="../../001.tleap_build/1ndv.wet.complex.prmtop"
coords="../../002.equilbration/09.equil"
MDINPUTS=(10.prod)
for input in ${MDINPUTS[@]}; do
$do_parallel -O -i ${input}.mdin -o ${input}.mdout -p $prmtop -c ${coords}.rst7 -ref ${coords}.rst7 -x ${input}.trj -inf ${input}.info -r
${input}.rst7
coords=$input
done
echo "Finished Production on `date` "
Now submit your job using:
sbatch prod.sh
Once production is finished you should see a "10.prod.mdout" and a "10.prod.trj". View the "10.prod.mdout" file to ensure that the simulation ran to completion.
Analysis
RMSD
The first analysis we will be performing is an RMSD analysis. This will tell us how much the ligand, protein, or complex moved from its initial position during simulation. We will also be creating a trajectory file with only the protein and ligand atoms. This will be useful if you wanted to view the trajectory in Chimera or another software.
To make the stripped trajectory and calculate RMSD we will be using a tool called CPPTRAJ. Create a file called cpptraj_strip_wat.in and add the following lines:
#!/usr/bin/sh parm ../001.tleap_build/1ndv.wet.complex.parm7 #read in trajectory trajin ../003.production/10.prod.trj #strip solvent strip :WAT:Na+:Cl- #create gas-phase trajectory trajout 1ndv_gas.trj nobox
Run this file using the following command:
cpptraj -i cpptraj_strip_wat.in
The .trj generating will be smaller and easier to load moving forward. Now lets create a new input file to calculate the RMSD. Create a file called cpptraj_rmsd.in and add the following lines:
#!/usr/bin/sh parm ../001.leap_build/1ndv.gas.complex.parm7 #trajin the trajectory trajin 1ndv_gas.trj #read in reference reference ../001.tleap_build/3wze.gas.complex.rst7 #compute RMSD (do not fit internal geometris first, included rigid body motions, convert frames to ns (framenum*.005) rmsd rms1 ":350&!(@H=)" nofit mass out 1ndv_lig_restrained_rmsd_nofit.dat time .005 #histogram the nofit rmsd histogram rms1,*,*,.1,* norm out 1ndv_lig_restrained_rmsd_nofit_histogram.dat rmsd rms2 ":1-349&!(@H=)" nofit mass out 1ndv_protein_restrained_rmsd_nofit.dat time .005 #histogram the nofit rmsd histogram rms1,*,*,.1,* norm out 1ndv_protein_restrained_rmsd_nofit_histogram.dat
Run the file using the command:
cpptraj -i cpptraj_rmsd.in
This file loads in our paramter file for the gas complex, followed by our trajectory and our reference of the first frame. It then computes the RMSD of just the ligand, and then just the protein as well as histogram data for them. These files can then be graphed.
Hydrogen Bonding
We will now analyze the hydrogen bond formed between the ligand and the protein. We will gain be using CPPTRAJ to do this. Note that this script will tell us all the hydrogen bonds that are formed and how long they persist in simulation, not just the ones between the ligand and the protein. Create a file called cpptraj_hbond.in and add the following lines:
#!/usr/bin/sh parm ../001.leap_build/1ndv.gas.complex.parm7 #trajin the trajectory trajin 1ndv_gas.trj #read in reference #wrap everything in one periodic cell - could cause problems, may comment out #autoimage if problems later autoimage #compute intra + water-mediated H-bonds hbond hb1 :1-350 out 1ndv_hbond.out avgout 1ndv_hbond_avg.dat solventdonor :WAT solventacceptor :WAT@-O nointramol bridgeout 1ndv_bridge-water.dat dist 3.0 angle 140 hbond hb2 :1-350 out 1ndv_sunitinib.hbond.out avgout 1ndv_sunitinib.hbond.avg.dat solventdonor :WAT solventacceptor :WAT nointramol bridgeout 1ndv_sunitinib.bridge-water.dat dist 3.0 angle 140
Run the script using the following command:
cpptraj -i cpptraj_hbond.in
MM-GBSA
The last analysis we will perform is an MM-GBSA calculation to estimate the free energy of binding. Create the file mmgbsa.in and add the following lines:
mmgbsa 1ndv analysis &general interval=1, netcdf=1, keep_files=0, / &gb igb=8 saltcon=0.0, surften=0.0072, surfoff=0.0, molsurf=0, / &nmode drms=0.001, maxcyc=10000, nminterval=250, nmendframe=2000, nmode_igb=1, /
This is a complex calcualtion and should not be run on the head node. Create a submissions script called mmgbsa.sh and add the following lines:
#! /bin/sh #SBATCH --job-name=production_unrest #SBATCH --ntasks-per-node=28 #SBATCH --nodes=1 #SBATCH --time=7-00:00:0 #SBATCH -p extended-28core cd $SLURM_SUBMIT_DIR
solv_parm="../001.tleap_build/1ndv.wet.complex.prmtop" complex_parm="../001.tleap_build/1ndv.gas.complex.parm7" receptor_parm="../001.tleap_build/1ndv.gas.receptor.parm7" lig_parm="../001.tleap_build/1ndv.gas.ligand.parm7" trajectory="../003.production/10_prod.trj" MMPBSA.py -O -i mmgbsa.in \
-o 1ndv_mmgbsa_results.dat \
-eo 1ndv_mmgbsa_perframe.dat \
-sp ${solv_parm} \
-cp ${complex_parm} \
-rp ${receptor_parm} \
-lp ${lig_parm} \
-y ${trajectory}