Difference between revisions of "2024 Denovo tutorial 1 with PDBID 2ITO"
Stonybrook (talk | contribs) (→Setting up the dummy atom) |
Stonybrook (talk | contribs) (→Setting up the dummy atom) |
||
| Line 35: | Line 35: | ||
Shown below here is the text of the pdb file which has already been modified: | Shown below here is the text of the pdb file which has already been modified: | ||
| − | [[File: | + | [[File: Dummypbd.png|thumb|center]] |
You can see at atom 23, which was supposed to be a carbon, has been changed the atom name and atom type to "Du". You can use that on your system too but please check in Chimera after editing to see if the atom has been marked purple. | You can see at atom 23, which was supposed to be a carbon, has been changed the atom name and atom type to "Du". You can use that on your system too but please check in Chimera after editing to see if the atom has been marked purple. | ||
Revision as of 09:15, 6 May 2024
Contents
Introduction
DeNovo Design aims to create a new ligand from scratch. It is incrementally designed bad added known structures of bioacitve compounds. The end result will hopefully produce a new ligand that can bind more tightly to a protein receptor than the current ligand can. There are three methods that will be introduced in this tutorial:
- Generic DeNovo Design
- Focused Fragment Design
- DeNovo Refinement
We will do denovo design with PDB: 2ITO.
Setting Up Your Environment
We will add more directories to the ones we used from the traditional virtual screening.
009_deNovoRefinement 010a_fragLib 010b_focusGrowth 010c_focusReScore 011_genericDeNovo
DeNovo Refinement
DeNovo Refinement can be used to assess the effects upon ligand and protein interactions by chaning a portion of the molecule. We can experiment with an interesting piece of the ligand and delete the remaining portion. This deleted portion needs to be replaced with a dummy atom. Dock will then find which fragments can be placed in this position that will have the least energy.
Setting up the dummy atom
A dummy atom would be required for setting up the deNovo design process. The dummy atom acts as a starting point for growing the molecule into a new molecule. For our 2TIO system, we would first open the file in Chimera:
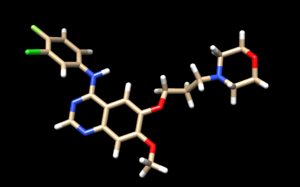
In this molecule, we will chose the ring chain to the right to cut it and replace with a dummy atom, since this is the group that falls deeper into the binding site of the protein. The molecule after changing into dummy would mark the dummy atom with a purple atom named Du as shown in Chimera:
We were able to change the file to dummy by first deleting the ring group in Chimera by selecting the group and continuously deleting the atoms of the group by Action > Atom > Delete. However we will still leave the atom point where we want to replace it into a dummy point. We will change that by opening the pdb file of the trimmed atom, replacing the name of the atom we want to change to Dummy by editing its name.
Shown below here is the text of the pdb file which has already been modified:
You can see at atom 23, which was supposed to be a carbon, has been changed the atom name and atom type to "Du". You can use that on your system too but please check in Chimera after editing to see if the atom has been marked purple.
Running DeNovo Refinement
Viewing New Molecules
Focused DeNovo Design
The subsequent two types of de novo design share a resemblance in which, with this approach, a specific structure is chosen to "anchor" the ligand to the protein. DOCK will then explore this structure throughout the protein to identify the optimal binding site. From this anchor point, a new ligand is developed. The distinction between focused and generic de novo design lies in the specific residues DOCK employs to build this new ligand.
Fragment Library Generation
To begin focus deNovo design, the first step is to construct a fragment library that is compatible with the ligand-complex system that one is interested in. In other to do so, we will use the original ligand along with the selected sphere from the surface step as a template for generating the fragment library and the algorithm to do so is embedded in DOCK. Thus, we will be gin by changing the working directory to the 013a.fragLib.in.
Here we will initiate an input file for the program:
vi fragLib.in
The following should be the input parameters for the file:
conformer_search_type flex write_fragment_libraries yes fragment_library_prefix 2tio_fragLib fragment_library_freq_cutoff 1 fragment_library_sort_method freq fragment_library_trans_origin no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.structure/2tio_ligand_hydrogens.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl ligand_outfile_prefix 2tio_focused write_orientations no num_scored_conformers 1 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
After finish compiling the input file for fragment generation, we can start running the program:
dock6 -i fragLib.in -o fragLib.out
The output file should be contain the following, note that if we fail to generate sufficient fragment library, there would not be enough input file for sequel steps:
- 2tio_focused_scored.mol2
- 2tio_fragLib_linker.mol2
- 2tio_fragLib_rigid.mol2
- 2tio_fragLib_scaffold.mol2
- 2tio_fragLib_sidechain.mol2
- 2tio_fragLib_torenv.dat
DeNovo Design
In this next step we will be navigating to the working directory for deNovo design using the Focus algorithm. The name of the directory for working would be 013b.foucusGrowth. As a sequel step from identifying the fragment library in the previous step, it would be required that you already achieved the library fragments set and store it correctly in the 013a.fragLib.
Initiating the input file with vi:
vi deNovoFocus.in
Insert the following content for the input:
conformer_search_type denovo dn_fraglib_scaffold_file ../013a.fragLib/2tio_fragLib_scaffold.mol2 dn_fraglib_linker_file ../013a.fragLib/2tio_fragLib_linker.mol2 dn_fraglib_sidechain_file ../013a.fragLib/2tio_fragLib_sidechain.mol2 dn_user_specified_anchor no dn_use_torenv_table yes dn_torenv_table ../013a.fragLib/2tio_fragLib_torenv.dat dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 1.0 dn_pruning_clustering_cutoff 100.0 dn_constraint_mol_wt 550.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 2 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees yes dn_output_prefix 2tio_focused use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
Start running the deNovo focus algorithm with the input as:
dock6 -i deNovoFocus.in -o deNovoFocus.out
After running the command, you should leave your program run until it executes. The program could not be run on server with mpi run. After you have successfully run the command, please check the directory output file to see if there have been any errors and the directory should contain multiple mol2 files in which the build.mol2 would be summarizing the molecules that have been built with denovo design algorithm.
ReScoring Designed Molecules
In other to make the multiples molecules suggested in the last step become more informative on which molecule would be the most likely to become the drug lead, in this step, the molecule set would be re-scored so that each would be compared with the binding of the original binding ligand to see if they would govern a similar effect to the original binder. The algorithm used for this scoring would be footprint similarity score, to begin, we would navigate to the directory designated for this section 013c.focusReScore. Here please initiate by creating the input file:
vi focusReScore.in
Insert the following content for the input:
conformer_search_type rigid
use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../013b.focusGrowth/2tio_focused.denovo_build.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary no footprint_similarity_score_secondary no pharmacophore_score_primary no pharmacophore_score_secondary no descriptor_score_primary yes descriptor_score_secondary no descriptor_use_grid_score no descriptor_use_multigrid_score no descriptor_use_continuous_score no descriptor_use_footprint_similarity yes descriptor_use_pharmacophore_score yes descriptor_use_tanimoto yes descriptor_use_hungarian yes descriptor_use_volume_overlap yes descriptor_fps_score_use_footprint_reference_mol2 yes descriptor_fps_score_footprint_reference_mol2_filename ../004.energy_min/2tio.lig.min_scored.mol2 descriptor_fps_score_foot_compare_type Euclidean descriptor_fps_score_normalize_foot no descriptor_fps_score_foot_comp_all_residue yes descriptor_fps_score_receptor_filename ../001.structure/2tio_protein_hydrogens_charges.mol2
descriptor_fps_score_vdw_att_exp 6
descriptor_fps_score_vdw_rep_exp 12 descriptor_fps_score_vdw_rep_rad_scale 1 descriptor_fps_score_use_distance_dependent_dielectric yes descriptor_fps_score_dielectric 4.0 descriptor_fps_score_vdw_fp_scale 1 descriptor_fps_score_es_fp_scale 1 descriptor_fps_score_hb_fp_scale 0 descriptor_fms_score_use_ref_mol2 yes descriptor_fms_score_ref_mol2_filename ../004.energy_min/2tio.lig.min_scored.mol2 descriptor_fms_score_write_reference_pharmacophore_mol2 no descriptor_fms_score_write_reference_pharmacophore_txt no descriptor_fms_score_write_candidate_pharmacophore no descriptor_fms_score_write_matched_pharmacophore no descriptor_fms_score_compare_type overlap descriptor_fms_score_full_match yes descriptor_fms_score_match_rate_weight 5.0 descriptor_fms_score_match_dist_cutoff 1.0 descriptor_fms_score_match_proj_cutoff 0.7071 descriptor_fms_score_max_score 20 descriptor_fingerprint_ref_filename ../004.energy_min/2tio.lig.min_scored.mol2 descriptor_hms_score_ref_filename ../004.energy_min/2tio.lig.min_scored.mol2 descriptor_hms_score_matching_coeff -5 descriptor_hms_score_rmsd_coeff 1 descriptor_volume_score_reference_mol2_filename ../004.energy_min/2tio.lig.min_scored.mol2 descriptor_volume_score_overlap_compute_method analytical descriptor_weight_fps_score 1 descriptor_weight_pharmacophore_score 1 descriptor_weight_fingerprint_tanimoto -1 descriptor_weight_hms_score 1 descriptor_weight_volume_overlap_score -1 gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl chem_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/chem.defn pharmacophore_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/ph4.defn
ligand_outfile_prefix 2tio_focus_rescore
write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Start running the rescore algorithm with the input as:
dock6 -i focusReScore.in -o focusReScore.out
If you have successfully computed the simulation you should receive the output results as followed: 4s0v_focus_rescore_footprint_scored.txt 4s0v_focus_rescore_hbond_scored.txt 4s0v_focus_rescore_scored.mol2
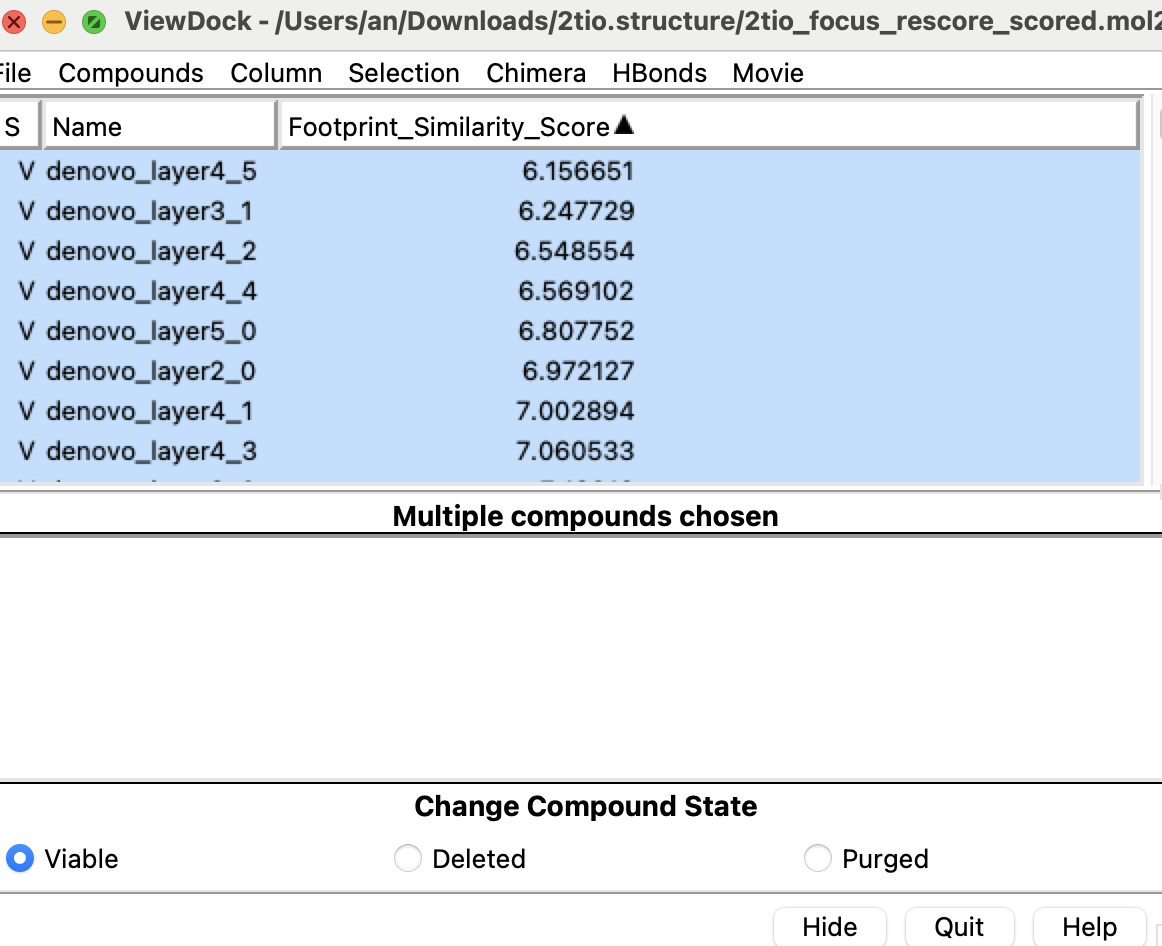
These are the sets of molecules suggested by deNovo being ranked based on the footprint scores (as mentioned above), and specifically with the hydrogen bonding scores. In other to view the results, you would need to load the result mol2 attached down to your local computer and view that with Chimera.
Below is loading the mol2 with ViewDock and ranking the file based on the footprint similarity score:
Generic DeNovo Design
For generic denovo design, we are not limiting DOCK to build new molecules using fragments present in the original ligand. Instead, we will be taking fragments from a fragment library.
Create the input file:
vi 2ito_generic.in
The file should contain these lines:
conformer_search_type denovo dn_fraglib_scaffold_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_scaffold.mol2 dn_fraglib_linker_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_linker.mol2 dn_fraglib_sidechain_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_sidechain.mol2 dn_user_specified_anchor no dn_torenv_table /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/fraglib_torenv.dat dn_name_identifier 2ito_generic dn_sampling_method graph dn_graph_max_picks 30 dn_graph_breadth 3 dn_graph_depth 2 dn_graph_temperature 100.0 dn_pruning_conformer_score_cutoff 100.0 dn_pruning_conformer_score_scaling_factor 2.0 dn_pruning_clustering_cutoff 100.0 dn_mol_wt_cutoff_type soft dn_upper_constraint_mol_wt 550.0 dn_lower_constraint_mol_wt 0.0 dn_mol_wt_std_dev 35.0 dn_constraint_rot_bon 15 dn_constraint_formal_charge 2.0 dn_heur_unmatched_num 1 dn_heur_matched_rmsd 2.0 dn_unique_anchors 1 dn_max_grow_layers 9 dn_max_root_size 25 dn_max_layer_size 25 dn_max_current_aps 5 dn_max_scaffolds_per_layer 1 dn_write_checkpoints yes dn_write_prune_dump no dn_write_orients no dn_write_growth_trees no dn_output_prefix 2ito_generic use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file /gpfs/projects/AMS536/2024/students/group_1_2ITO/002_surface_spheres/2ITO.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no grid_score_primary yes grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix /gpfs/projects/AMS536/2024/students/group_1_2ITO/003_gridbox/grid minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 250 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/vdw_de_novo.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.10/parameters/flex_drive.tbl
Create a slurm file to run this:
#!/bin/bash #SBATCH --job-name=2ito_generic #SBATCH --ntasks=28 #SBATCH --nodes=1 #SBATCH --time=48:00:00 #SBATCH -p long-28core dock6 -i 2ito_generic.in
After the program has run, you can use the following command to see how many ligands has been generated. The program has made 368 potential ligands.
grep MOLECULE 2ito_generic.denovo_build.mol2 | wc -l
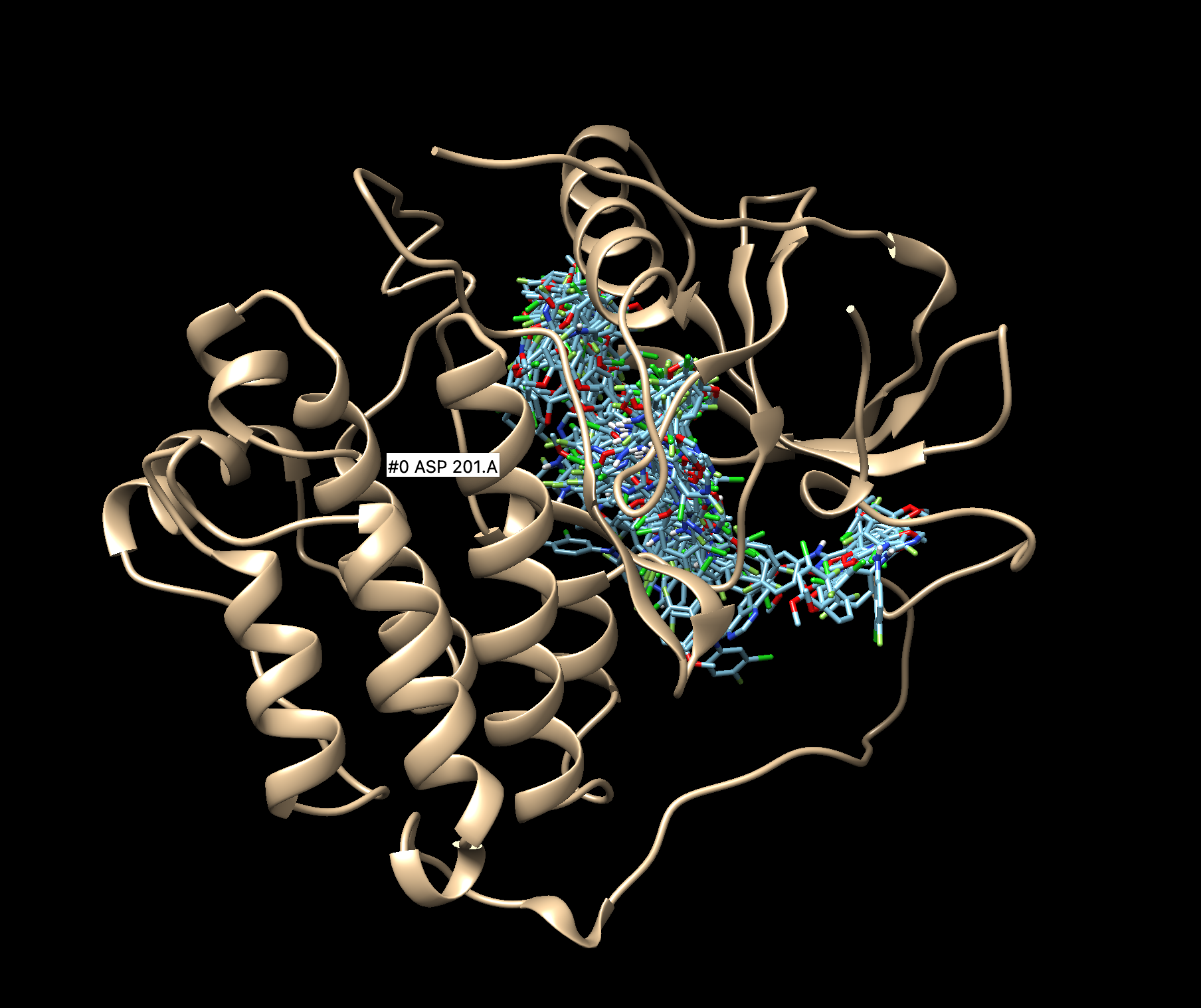
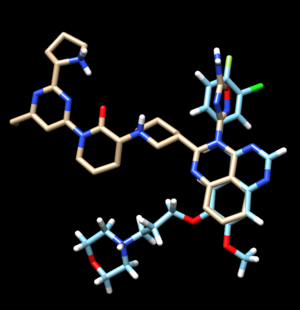
The output file we are going to move to our local computer is 2ito_generic.denovo_build.mol2. We can see the ligands the program has created.
Go into Chimera and open the molecule on ViewDock. (Tools >> Surface/Binding Analysis >> ViewDock)
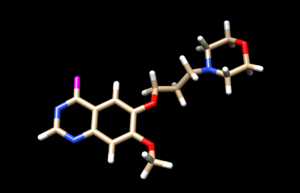
Use Column >> Show >> Grid_Score to find the ligand with the lowest energy. The ligand with the lowest score has a score of -93.318.
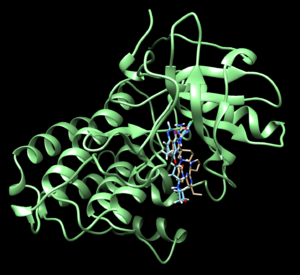
We can compare this with the energy minimized ligand and see how it is positioned differently compared to the generated ligand (denovo_designed ligand is brown, energy minimized ligand is blue). This original energy minimized ligand has a grid score of -71.423.
We can also observe how the designed ligand and the original ligand interact with the receptor.