Difference between revisions of "2009 DOCK tutorial with neuraminidase"
(→Manipulating the PDB file using vi for DOCK) |
Stonybrook (talk | contribs) (→Preparing the Enzyme and Ligand in Chimera) |
||
| Line 112: | Line 112: | ||
on the command line, type "delete selected" | on the command line, type "delete selected" | ||
| + | To prepare the ligand: | ||
| − | + | go to Tools -> Structure Editing -> Dock Prep. Continue on with the selected by default. After pressing OK, use the AM1-BCC charge method. | |
| + | Save the molecule in .mol2 format as 'chargedligand.mol2' | ||
| + | |||
| + | Alternatively try the following: | ||
*add hydrogens using '''Tools → Structure Editing → Add H'''. Again, select '''Unspecified determined by method''' for the histidine protonation. | *add hydrogens using '''Tools → Structure Editing → Add H'''. Again, select '''Unspecified determined by method''' for the histidine protonation. | ||
Revision as of 19:12, 9 June 2009
Contents
- 1 About DOCK
- 2 Making sure DOCK in your PATH
- 3 About Neuraminidase
- 4 Downloading the PDB complex (1F8B)
- 5 Manipulating the PDB file using vi for DOCK
- 6 Preparing the Enzyme and Ligand in Chimera
- 7 Generation of Enzyme Surface, Spheres, and Grid for DOCK
- 8 DOCKing
- 9 Using a .csh File to Generate Enzyme Surface, Spheres, and Grid for DOCK and to run DOCK with a Single Command
- 10 Docking Results
About DOCK
DOCK was developed by Irwin D. "Tack" Kuntz, Jr., PhD and colleagues at UCSF. Please see the webpage at UCSF DOCK.
DOCK is a molecular docking program used in drug discovery. This program, given a protein active site and a small molecule, tries to predict the correct binding mode of the small molecule in the active site, and the associated binding energy. Small molecules with highly favorable binding energies could be new drug leads. This makes DOCK a valuable drug discovery tool. DOCK is typically used to screen massive libraries of millions of compounds against a protein to isolate potential drug leads. These leads are then further studied, and could eventually result in a new, marketable drug.
In the following tutorial, commands to be entered at the terminal and command line of Chimera will be in dotted boxes, like this one:
A dotted box
Whether at terminal or command line will be distinguished by text
Programs central to DOCK are:
SPHGEN- This program generates sets of overlapping spheres to describe the shape of a molecule or molecular surface. It identifies the active site and other sites of interest and generates spheres that fill that site.
GRID - This program generates the grid files necessary for rapid score evaluation in DOCK (rapid scoring evaluation can be done using contact or energy scoring).
DOCK - This program matches the spheres generated by SPHGEN with ligand atoms and use scoring grids which are previously calculated using GRID to evaluate the ligand orientations
Making sure DOCK in your PATH
Try the command
which dock6
If dock6 is not in your path, edit your .cshrc (or equivalent) to include the dock6/bin folder in your PATH.
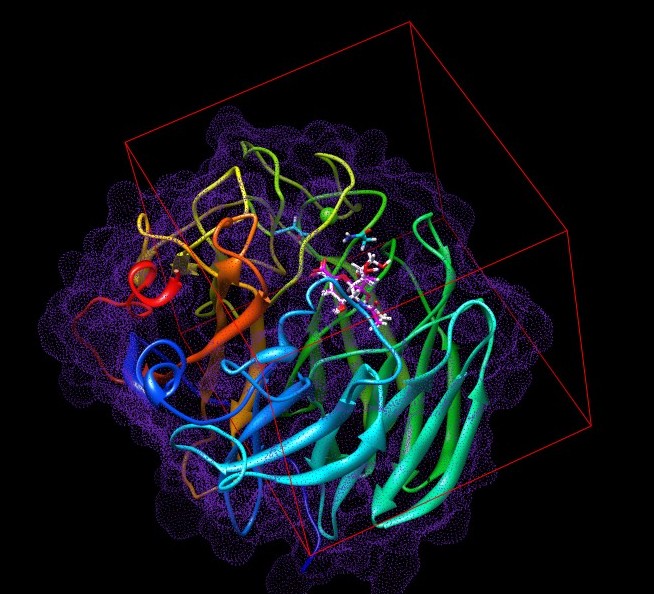
About Neuraminidase
Neuraminidase is an enzyme family which cleaves glycosidic linkages. In the process of Influenzavirus infection, viral Neuraminidase helps newly assembled virus to be released from host cells by hydrolyzing glycosidic bonds which connect the infected host cells and the hemagglutinin on the surface of progeny viruses.
Downloading the PDB complex (1F8B)
Download the neuraminidase file from here into your working directory. To get the monomer, download using the blue file arrow icon. Choose biological unit gz under download files folder at the left side. In that way you can downlad a biological gz file. If you download it by using "fetch by ID" in Chimera, you can only download a pdb gz file.
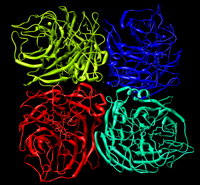
If the pdb gz file is downloaded the monomeric structure of neuraminidase with the bound ligand is seen.
However if the biological gz file is downloaded a tetramer complex of neuraminidase is seen.
When docking, we will only choose a monomer of the whole protein and its ligands instead of the whole tetramer. And it will be painful if you follow the procedure below, use vi to prepare the enzyme and ligand. Because you need to make sure the ligand you extract from the whole pdb files is right the one that match the monomer you've chosen.
Manipulating the PDB file using vi for DOCK
The PDB file can be opened and manipulated in the text editor vi.
For example it is very easy to isolate a certain ligand and create a new pdb file without having to work through Chimera. This alternate method may be appropriate in certain cases when the pdb file was not carefully compiled resulting in odd grouping of certain molecules and atoms. It is best to use vi editing in conjunction with Chimera to achieve greatest efficiency.
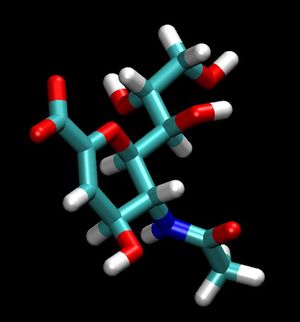
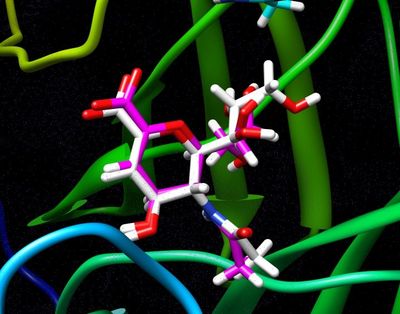
For a simple example, we can isolate the ligand 2-Deoxy-2,3-dehydro-N-acetyl-neuraminic Acid (tagged DAN in this pdb), by simply copy a few lines out of the original pdb and pasting them into a new one. To do this simply open up the original pdb file:
vi 1F8B.pdb
Find the ligand and copy it into a separate pdb file.
The ligand can now be easily prepared for DOCK by opening it with Chimera and using its built in DOCK prep program.
Many other functions such as grouping ligands and deleting atoms or residues may be done in a facile manner by simply editing the pdb file through the vi text editor.
Preparing the Enzyme and Ligand in Chimera
Open the pdb file in chimera using Open or Fetch by ID from the File menu.To visualize the protein and the ligand, click Presets → Interactive 1 (ribbons). To access the command line, go to Tools → General Controls → Command Line.
Before you begin make sure to delete the extra molecules from the pdb file. To do so, in chimera open the 1F8B.pdb file. Select->Residue->HOH in the command line type
delete selected
repeat this for MAN and NAG as well.
Preparing the enzyme
First you want to delete the ligand and prepare the enzyme for DOCK by charging it.
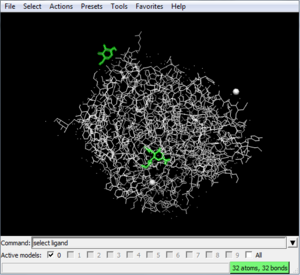
To delete the ligand: In the command line of Chimera, type
select ligand
to select the ligand. To delete the ligand, type:
delete ligand
Or alternatively, you can use the chimera tool bar directly.
To charge the enzyme:
- go to Tools → Structure Editing → Dock Prep
- In the first dialogue box that appears select ok without making changes. In the second, select Unspecified determined by method for the histidine protonation then ok.
- You will then get a dialogue box asking you to enter the net charges of specific molecules. Hit ok to run without changing the calculated net charges.
- Save the .mol2 file as nacharged.mol2. This is your charged enzyme.
We will use the 'dms' program to generate the surface of the enzyme. In order to use this program, the hydrogens on the enzyme must be removed. this pdb file is already stripped of H, but to do this:
- go to Select → Chemistry → Element → H
- then hit Actions → Atoms/Bonds → Delete
- Save the charged receptor in .pdb format by File → Save pdb name it naforDOCK.pdb
For more details about DockPrep, use http://www.cgl.ucsf.edu/chimera/1.2199/docs/ContributedSoftware/dockprep/dockprep.html File:Example.jpg
One need to notice that the structure downloaded form PDB usually contains substrate, ligands, ions cofactors and water etc. While when preparing enzyme for docking, we should only keep substrate and ions.
Preparing the Ligand
Now you want to focus on just the ligand, so you need to delete the enzyme.
Open a new session in chimera using the original '1f8b.pdb'
To delete the enzyme, in the command line type:
select ligand select invert all models delete selected
However, after this , NAG(which is not a ligand is still left), maybe because the Chimera is not clever enough, so you need to remove NAG manually:
on the tool bar, click "select->residue->NAG" on the command line, type "delete selected"
To prepare the ligand:
go to Tools -> Structure Editing -> Dock Prep. Continue on with the selected by default. After pressing OK, use the AM1-BCC charge method. Save the molecule in .mol2 format as 'chargedligand.mol2'
Alternatively try the following:
- add hydrogens using Tools → Structure Editing → Add H. Again, select Unspecified determined by method for the histidine protonation.
- add charge using Tools → Structure Editing → Add Charge. Choose the AMBER ff03.r1 charge model that is selected by default. After pressing OK, use the AM1-BCC charge method. Unlike prior steps, this may take several minutes.
- Save the molecule in .mol2 format as 'chargedligand.mol2'
Alternatively, one can just apply dockprep to get the mol2 file of the charged ligand.
For reference the total charge on the ligand DAN will be -1.
Generation of Enzyme Surface, Spheres, and Grid for DOCK
Enzyme Surface
To generate the enzyme surface, you use the enzyme stripped of hydrogens (this is your naforDOCK.pdb file that we generated previously) in the dms program.
the dms program is found at: /home/sudipto/dms_program
In terminal (you must be in the directory where you saved naforDOCK.pdb):
/home/sudipto/dms_program naforDOCK.pdb -n -w 1.4 -v -o nasurface.ms
These commands specify the dms program for the naforDOCK file, -n calculates normals for surface points, -w changes probe radius to 1.4, -v verbose, and -o specifies the output file name (in your case nasurface.ms).
Spheres
To generate spheres, you will use a program called sphgen. Before using sphgen, you need to create a INSPH file.
To create an INSPH file, in terminal:
vi INSPH
this will create a file called INSPH
type in
nasurface.ms R X 0.0 4.0 1.4 naspheres.sph
when sphgen runs this, it will use the .ms surface file you just created using dms and give an output file of naforDOCK.sph
to run type in:
sphgen -i INSPH -o OUTSPH
it would take several minutes
An example of new created OUTSPH file
density type = X reading nasurface.ms type R # of atoms = 3067 # of surf pts = 81632 finding spheres for nasurface.ms dotlim = 0.000 radmax = 4.000 Minimum radius of acceptable spheres? 1.39999998 output to naspheres.sph clustering is complete 53 clusters
NOTE:If the output file OUTSPH or receptor.sph or any of the temp files (temp1.ms, temp2.sph or temp3.atc) exist, a core dump will occur and sphgen will fail. Delete all these files before running sphgen.
now to isolate only spheres surrounding the ligand, you will use sphere_selector
type in:
sphere_selector naspheres.sph chargedligand.mol2 10.0
To visualize the spheres surrounding the ligand that you have just created, you need to convert the spheres into pdb format. You will use the showsphere program to do this.
type in:
showsphere
when prompted type in:
Enter name of sphere cluster file: naspheres.sph Enter cluster number to process (<0 = all): 53 Generate surfaces as well as pdb files (<N>/Y)? Y Enter name for output PDB file name: naligandspheres.pdb Enter name for output surface file name: Any name you want.
You can use Chimera to open nasurface.ms to visualize the surface.
Grid
The last files you need to generate before you can run DOCK are grid files. The grid is the energy map that allows scoring of the DOCK performed. To run grid you need to create 2 input files: showbox.in and grid.in using vi.
In terminal type
vi showbox.in
or
vi grid.in
and create the following files
showbox.in
Y 5.0 naspheres.sph 1 nabox.pdb
grid.in
compute_grids yes grid_spacing 0.5 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file nacharged.mol2 box_file nabox.pdb vdw_definition_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
For the sake of time, your grid spacing is 0.5 A. For an actual experiment, there should be less space between the grid points (0.3A is recommended). Also, we have weakened the van der Waals interactions from 6-12 down to 6-9 in our attractive_exponent and repulsive_exponent parameters. Be aware of these facts when going to set up your own DOCK experiment. This makes the repulsive
now you are ready to run showbox and grid. In terminal type:
showbox < showbox.in
to generate a grid box. To generate a grid, type:
grid -i grid.in
It would take 20-40 minutes. Have a cup of tea. You could also add '-o grid.out &' to save the grid progress statistics to a file instead and have the job run in the background while you do other things.
You have created two output files: grid.nrg and grid.bmp. You are now ready to run DOCK. Make sure these files exist. The grid.nrg contains the energy grid and should be several MB.
DOCKing
DOCK can be done with a flexible ligand or a rigid ligand. You choose which way you would like to run the DOCK depending on your system. Either way, you will need to create an input file.
For a rigid ligand, your input file is rigid.in which you will create using vi.
In terminal, type
vi rigid.in
the rigid.in file is:
ligand_atom_file chargedligand.mol2 ligand_outfile_prefix rigid limit_max_ligands no read_mol_solvation no write_orientations no write_conformations no skip_molecule no calculate_rmsd yes use_rmsd_reference_mol no rank_ligands no num_scored_conformers_written 1 orient_ligand yes automated_matching yes receptor_site_file naspheres.sph max_orientations 500 critical_points no chemical_matching no use_ligand_spheres no flexible_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid dock3.5_score_secondary no continuous_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_final_min_add_internal no simplex_random_seed 0 atom_model all vdw_defn_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /home/rizzo/AMS536software/dock6/parameters/flex.defn flex_drive_file /home/rizzo/AMS536software/dock6/parameters/flex_drive.tbl
Now to perform rigid docking type in the command line
dock6 -i rigid.in -o rigid.out
For flexible docking, first create the file flex.in using vi
vi flex.in
which will contain the following details
ligand_atom_file ./chargedligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol no orient_ligand yes automated_matching yes receptor_site_file ./selected_spheres.sph max_orientations 500 critical_points no chemical_matching no use_ligand_spheres no flexible_ligand yes min_anchor_size 40 pruning_use_clustering yes pruning_max_orients 100 pruning_clustering_cutoff 100 use_internal_energy yes internal_energy_att_exp 6 internal_energy_rep_exp 12 internal_energy_dielectric 4.0 use_clash_overlap no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid dock3.5_score_secondary no continuous_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_final_min no simplex_random_seed 0 atom_model all vdw_defn_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /home/rizzo/AMS536software/dock6/parameters/flex.defn flex_drive_file /home/rizzo/AMS536software/dock6/parameters/flex_drive.tbl ligand_outfile_prefix flex write_orientations no num_scored_conformers 50 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
The command to do flexible docking is
dock6 -i flex.in -o flex.out
Using a .csh File to Generate Enzyme Surface, Spheres, and Grid for DOCK and to run DOCK with a Single Command
The above tells you how to step by step go through and create all files necessary for DOCK. Each program uses a file generated by the program run before it. It is also possible to create a single .csh file which specifies all of the programs and files to use so that all you will need to do is type "compute" and the computer will go through all of the programs for you.
First you will need to create certain files that the programs need. These are the same files that you created if you followed the step by step protocol (although right now some files still have different names and are not all there yet).
Files needed
showsph.in
naspheres.sph 1 N naligandspheres.pdb
INSPH
nasurface.ms R X 0.0 4.0 1.4 naspheres.sph
showbox.in
Y 5.0 selected_spheres.sph 1 rec_box.pdb
grid.in
compute_grids yes grid_spacing 0.5 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4 bump_filter yes bump_overlap 0.75 receptor_file nacharged.mol2 box_file rec_box.pdb vdw_definition_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
rigid.in
ligand_atom_file ./chargedligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol no orient_ligand yes automated_matching yes receptor_site_file ./selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no flexible_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid dock3.5_score_secondary no continuous_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_final_min no simplex_random_seed 0 atom_model all vdw_defn_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /home/rizzo/AMS536software/dock6/parameters/flex.defn flex_drive_file /home/rizzo/AMS536software/dock6/parameters/flex_drive.tbl ligand_outfile_prefix rigid write_orientations no num_scored_conformers 10 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
flex.in
ligand_atom_file ./chargedligand.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol no orient_ligand yes automated_matching yes receptor_site_file ./selected_spheres.sph max_orientations 500 critical_points no chemical_matching no use_ligand_spheres no flexible_ligand yes min_anchor_size 40 pruning_use_clustering yes pruning_max_orients 100 pruning_clustering_cutoff 100 use_internal_energy yes internal_energy_att_exp 6 internal_energy_rep_exp 12 internal_energy_dielectric 4.0 use_clash_overlap no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix grid dock3.5_score_secondary no continuous_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_final_min no simplex_random_seed 0 atom_model all vdw_defn_file /home/rizzo/AMS536software/dock6/parameters/vdw_AMBER_parm99.defn flex_defn_file /home/rizzo/AMS536software/dock6/parameters/flex.defn flex_drive_file /home/rizzo/AMS536software/dock6/parameters/flex_drive.tbl ligand_outfile_prefix flex write_orientations no num_scored_conformers 50 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Now you can create the script files, prep.csh, rigid.csh, and flex.csh. Run prep.csh to generate sphere and grid information for use in docking. Run rigid.csh to perform rigid docking and flex.csh to perform flexible docking.
prep.csh
#!/bin/csh echo Setting up #setup set dock6bin = /opt/software/AMS536software/dock6/bin set workdir = /home/rizzo/dock_project mkdir -p $workdir cd $workdir echo Generating the molecular surface ~sudipto/dms_program naforDOCK.pdb -n -w 1.4 -v -o nasurface.ms echo Generating spheres #delete temp files rm OUTSPH naspheres.sph temp1.ms temp2.sph temp3.atc #generate spheres $dock6bin/sphgen -i INSPH -o OUTSPH #select spheres near binding site $dock6bin/sphere_selector naspheres.sph chargedligand.mol2 10.0 $dock6bin/showsphere < showsph.in echo Generating grid #generate grid box $dock6bin/showbox < showbox.in #generate grid $dock6bin/grid -i grid.in
rigid.csh
#!/bin/csh echo Setting up #setup set dock6bin = /opt/software/AMS536software/dock6/bin set workdir = /home/rizzo/dock_project mkdir -p $workdir cd $workdir echo Performing rigid docking! #perform rigid docking $dock6bin/dock6 -i rigid.in -o rigid.out
flex.csh
#!/bin/csh echo Setting up #setup set dock6bin = /opt/software/AMS536software/dock6/bin set workdir = /home/rizzo/dock_project mkdir -p $workdir cd $workdir echo Performing flexible docking! #perform flexible docking $dock6bin/dock6 -i flex.in -o flex.out
*Note: In all these scripts, you will not be able to modify Professor Rizzo's directory, so at the line
set workdir = /home/rizzo/dock_project
replace rizzo with your directory.
Summarily, to perform a complete rigid and flexible docking, type:
./prep.csh ./rigid.csh ./flex.csh
When the process is complete, you should end with .out files detailing the calculation of your DOCKed ligand, as well as rigid_scored.mol2 and flex_scored.mol2 files that contain topology and coordination information about the best scored ligand for each method.