Difference between revisions of "MD Simulation: Protein in Water"
Stonybrook (talk | contribs) |
Stonybrook (talk | contribs) |
||
| Line 173: | Line 173: | ||
The final step before simulation is to add enough ions to the system to neutralize the net charge or, alternatively, add enough ions to neutralize the net charge <i>and</i> reach some physiological concentration. If you are simulating a system in an attempt to replicate some experimental observable, for example, it is important to use the same salt concentration in your system as is used in the experiment. A standard value for salt concentration often used to replicate human physiology is 100 mM. | The final step before simulation is to add enough ions to the system to neutralize the net charge or, alternatively, add enough ions to neutralize the net charge <i>and</i> reach some physiological concentration. If you are simulating a system in an attempt to replicate some experimental observable, for example, it is important to use the same salt concentration in your system as is used in the experiment. A standard value for salt concentration often used to replicate human physiology is 100 mM. | ||
| + | The 'genion' tool searches through your coordinate file and will randomly replace water molecules with ions. In order for it to work, however, it requires what a pre-processed input file (with extension .tpr) that contains all of the information from both the coordinate and topology files. In order to generate such a file, use the tool 'grompp'. Aside from the coordinate and topology file, 'grompp' also requires that you provide a MD paramter file (.mdp) as input. Because the .tpr file we are generating is not going to be used for dynamics, rather it is just going to be used to add ions, the contents of the .mdp file are not important. Nevertheless, it still must be provided on the command line. An example .mdp file with all of the default parameters can be found [[here]]. | ||
| + | grompp -f genion.mdp -c protein_sol.gro -p topol.top -o genion_input.tpr | ||
| + | The 'gr | ||
Revision as of 13:44, 21 July 2011
Contents
Purpose
The purpose of this tutorial is to provide an introduction to the fundamental commands needed to set up, run, and analyze MD simulations in GROMACS. This tutorial assumes you have already correctly installed GROMACS. This tutorial was written using GROMACS 4.5.4.
Download and Prepare PDB File
Ubiquitin is a small and perfectly ordinary protein, making it ideal for an introductory tutorial. A crystal structure can be found at the PDB website under the accession code 1UBQ. Before proceeding to the next step, now is a good time to use your favorite text editor to check for a few things:
1.) Are crystal waters present in the PDB file? In the case of 1UBQ, there are many crystal waters. Ubiquitin is non-enzymatic, though, so the waters are not important for an active site mechanism, for example. It is a good idea to delete the waters now and allow the GROMACS solvation tool to fill them back in later.
2.) Are there any ligands or non-standard residues present in the PDB file? In the case of 1UBQ, there are no ligands or non-standard residues. Ligand preparation and inclusion is covered in another tutorial (MD Simulation: Protein-Ligand Complex). Without going into too much detail, non-standard residues are okay so long as the residue name and atom names conform to the corresponding entry in the residue topology file (.rtp) of the force field you choose.
3.) Are there any residues with missing atoms in the PDB file? In the case of 1UBQ, there are not. If there were, however, you would need to take extra preparation steps beforehand to fix the broken residues or, if they are at one of the protein termini, consider removing them from the file.
GROMACS Coordinate and Topology Files
The first step in MD simulation with GROMACS is to create GROMACS-compatible coordinate (.gro) and topology (.top) files. Strictly speaking, .pdb files are GROMACS-compatible, but we might as well convert to a .gro coordinate file to keep in line with the spirit of the tutorial. The command line execution looks like this:
pdb2gmx -f 1UBQ.pdb -o protein.gro -p topol.top -ignh -v
When prompted, choose '1' for the AMBER03 force field, and '1' again for the TIP3P water model. These selections are fine for this tutorial, but make sure you think very carefully about your choice before picking a force field in your research. You may ask yourself, should I use an all-atom force field or a united-atom force field? Is my force field selection compatible with lipids or small molecules that I want to use? Can I compare the results of simulations using my force field to other simulations I performed previously / found in the literature? etc.
After you have picked the force field and a solvent model compatible with that force field, it is a good idea to read through the output on the screen to make sure there are no Errors or Warnings. In this case, it looks like there actually was one Warning:
WARNING: there were 0 atoms with zero occupancy and 28 atoms with
occupancy unequal to one (out of 602 atoms). Check your pdb file.
A quick look back at the original PDB file reveals that the four residues residues at the C-terminus of ubiquitin in 1UBQ were not as well resolved as the rest of the protein. Pdb2gmx noticed that, too, and adjusted the occupancy of each to '1'. This should be okay for now, as the atomic positions of these residues will be refined later in the minimization and equilibration steps. Aside from that one warning, it appears pdb2gmx changed a few residue names and atom names to conform to the names used in the AMBER03 .rtp file (found within ~/share/gromacs/top/amber03.ff/ of your local GROMACS directory). Ensure that the changes make sense, and it is okay to proceed.
There are three output files resulting from pdb2gmx:
1.) protein.gro # gromacs-format coordinate file 2.) topol.top # gromacs-format topology file 3.) posre.itp # position restraint file
It is very important to know and understand the contents of each file before continuing. Make sure you look through each file until you are able to make sense of the information contained within each.
Coordinate file
The GROMACS coordinate file contains the following:
Ubiquitin from 1UBQ - no water molecules (yet)
1231
1MET N 1 2.734 2.443 0.261
1MET H1 2 2.807 2.485 0.207
1MET H2 3 2.771 2.413 0.349
...
76GLY C 1229 4.003 3.999 3.543
76GLY OC1 1230 4.086 3.957 3.625
76GLY OC2 1231 3.893 4.052 3.569
3.06857 3.23738 3.66170
GROMACS coordinate files always contain three special lines. The first line is a title - it is good practice to use a detailed title specific to the system being simulated. The second line is the number of atoms. The final line contains the box vectors (in nanometers). The lines in between contain the residue number, residue name, atom name, atom number, and cartesian coordinates (in nanometers) for each atom in the system.
Topology file
The GROMACS topology file contains the following sections:
#include "amber03.ff/forcefield.itp"
[ moleculetype ] Protein_chain_A 3 [ atoms ] ... [ bonds ] ... [ pairs ] ... [ angles ] ... [ dihedrals ] ... [ dihedrals ] ... #ifdef POSRES #include "posre.itp" #endif
[ system ] Ubiquitin [ molecules ] Protein_chain_A 1
The first section is an include statement that, when this file is processed, pastes the bonded and non-bonded information specific to your force field directly into the topology file. It is a good idea to familiarize yourself with the contents of the 'forcefield.itp' file, and the files included within it, all found under ~/share/gromacs/top/amber03.ff/.
The second section (usually) contains 6 or 7 sub-sections. First, under '[ moleculetype ]' is the name of the molecule followed by a number. In this case, it is by default called 'Protein_chain_A'. The number N indicates that for this molecule, exclude non-bonded interactions of all bonded neighbors up to N bonds away. Unless you have a very good idea of what you are doing, you should not change this number. A given moleculetype is followed by (and it must be in this order) atom information, bond information, pair-wise exclusion information, angles information, dihedral information, improper dihedral information, and, optionally, position restraint information. Not all of the sections must be present. Further, this entire section is repeated for each type of molecule that you have. For example, if you are simulating a protein in water, you will have two consecutive '[ moleculetype ]' sections.
The final section only includes two sub-sections. Under '[ system ]' is a system title chosen by the user, and under '[ molecules ]' is a list of the moleculetypes found in the topology file, followed by the number of times each moleculetype appears in the coordinate file. For example if you have a coordinate file that contains a protein in 10,000 water molecules, your topology file should look roughly like:
#include "amber03.ff/forcefield.itp"
[ moleculetype ] Protein 3 ...
[ moleculetype ] SOL 1 ...
[ system ] Protein in water [ molecules ] Protein 1 SOL 10000
where the moleculetypes under '[ molecules ]' are listed in the same order as they appear in the topology file. Note that the topology file you generated already contains an #include statement to include the moleculetype for TIP3P water and for ions, but because there are no water molecules or ions in our coordinate file, they are not yet listed under '[ molecules ]'. Chapter 5 of the GROMACS manual is an excellent resource for further information regarding molecular topologies.
Position restraints file
The final file that was created by pdb2gmx is the position restraints file:
[ position_restraints ]
1 1 1000 1000 1000
5 1 1000 1000 1000
7 1 1000 1000 1000
...
Each line indicates an atom number, a functional form (1 = harmonic restraint), and force constants in cartesian space. By default, only the protein backbone atoms are listed in this file. Looking back at the topology file, you can see that if 'POSRES' is defined when you begin simulation, then these parameters will be included in the molecular topology, thus restraining the backbone of the protein during simulation. If 'POSRES' is not defined, then the position restraints are ignored.
Box Preparation
In the next few steps, you will define a box size for your system, center the protein in the box, solvate, and finally, add counterions to the system. The files you need to start this step are:
1.) protein.gro # gromacs-format coordinate file 2.) topol.top # gromacs-format topology file 3.) posre.itp # position restraint file
editconf
The GROMACS tool editconf is very useful to change the format of your coordinate files, to rotate and translate coordinate files, to define the box size, among other things. Typing 'editconf -h' will display a brief description of its capabilities. To create a rectangular box around the protein, type:
editconf -f protein.gro -o protein_box.gro -bt triclinic -d 1.2 -c
The only input needed is the coordinate file generated previously. With '-bt triclinic' you are choosing to create a rectangular box. It is important to note that rectangular boxes can be very inefficient, especially for globular proteins like ubiquitin. In this tutorial, we will keep with the rectangular box, but in the future, I would highly recommend using '-bt dodecahedron' for globular proteins. The option '-d 1.2' creates a buffer of 1.2 nm between the outside of the protein and the edge of the box, and '-c' centers the protein in the box and puts the corner of the box at {0, 0, 0} in cartesian space. The output file is called 'protein_box.gro'.
The most important question to ask now is why choose 1.2 nm? The short answer is that you don't want the protein to 'see' its periodic image across the boundary of the box. If you refer to the original paper for the AMBER 03 force field (Duan, et al., J. Comput. Chem. 2003, 24, 1999-2012), they use short-range VDW and electrostatic cut-offs of 0.8 nm - when you begin simulating, it is very important to do the same. In order for the protein to avoid seeing its image across the periodic boundary, it must be at least twice the cut-off distance from the next nearest image of itself. That being said, the space between the protein and the edge of the box only really needs to be 0.8 nm, but it is still a good idea to include extra space especially if there is a chance the protein might unfold a little bit or if is an oblong shape. It is better to have a slightly larger box size now than to find out later that your protein was interacting with its periodic image during the simulation. (See the image below).
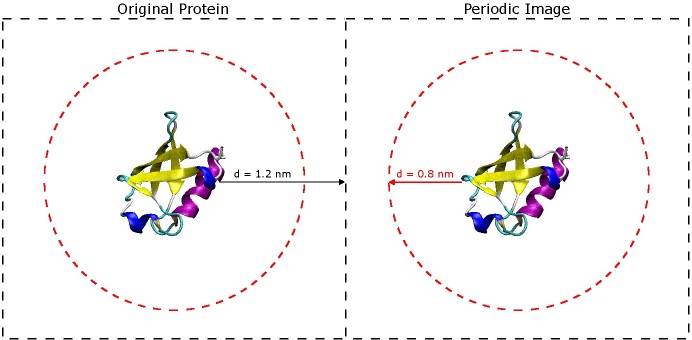 The dashed black squares indicate the box boundaries, and the dashed red circles indicate the short-range VDW and electrostatic cut-offs. So long as the red circles do not overlap, the protein will not be able to "see" its periodic image.
The dashed black squares indicate the box boundaries, and the dashed red circles indicate the short-range VDW and electrostatic cut-offs. So long as the red circles do not overlap, the protein will not be able to "see" its periodic image.
genbox
The next GROMACS command, 'genbox', will fill the box with solvent molecules.
genbox -cp protein_box.gro -cs spc216.gro -o protein_sol.gro -p topol.top
The '-cp' flag specifies the coordinate file of the system to be solvated, and '-cs spc216.gro' tells genbox to use an SPC (a simple three-point water model) coordinate file to populate the box. Thee output coordinate file is called 'protein_sol.gro'. The topology file, 'topol.top', is also provided on the command line. If you check the end of the topology file, you can see that it has been updated in the following way:
[ molecules ] Protein_chain_A 1 SOL 5760
At this point, it is useful to open 'protein_sol.gro' in VMD to ensure that the protein is correctly placed in the center of the box and there are no strange solvent artifacts.
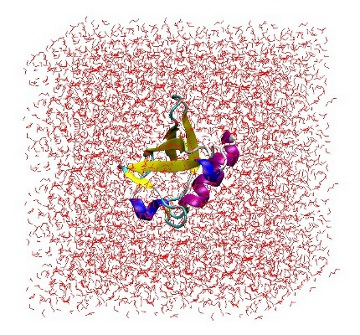 1UBQ in a triclinic box solvated with 5,760 water molecules.
1UBQ in a triclinic box solvated with 5,760 water molecules.
genion
The final step before simulation is to add enough ions to the system to neutralize the net charge or, alternatively, add enough ions to neutralize the net charge and reach some physiological concentration. If you are simulating a system in an attempt to replicate some experimental observable, for example, it is important to use the same salt concentration in your system as is used in the experiment. A standard value for salt concentration often used to replicate human physiology is 100 mM.
The 'genion' tool searches through your coordinate file and will randomly replace water molecules with ions. In order for it to work, however, it requires what a pre-processed input file (with extension .tpr) that contains all of the information from both the coordinate and topology files. In order to generate such a file, use the tool 'grompp'. Aside from the coordinate and topology file, 'grompp' also requires that you provide a MD paramter file (.mdp) as input. Because the .tpr file we are generating is not going to be used for dynamics, rather it is just going to be used to add ions, the contents of the .mdp file are not important. Nevertheless, it still must be provided on the command line. An example .mdp file with all of the default parameters can be found here.
grompp -f genion.mdp -c protein_sol.gro -p topol.top -o genion_input.tpr
The 'gr