Difference between revisions of "MD Simulation: Protein in Water"
Stonybrook (talk | contribs) (→Analysis) |
Stonybrook (talk | contribs) |
||
| Line 25: | Line 25: | ||
occupancy unequal to one (out of 602 atoms). Check your pdb file. | occupancy unequal to one (out of 602 atoms). Check your pdb file. | ||
| − | A quick look back at the original PDB file reveals that the four | + | A quick look back at the original PDB file reveals that the four residues at the C-terminus of ubiquitin in 1UBQ were not as well resolved as the rest of the protein. Pdb2gmx noticed that, too, and adjusted the occupancy of each to '1'. This should be okay for now, as the atomic positions of these residues will be refined later in the minimization and equilibration steps. Aside from that one warning, it appears pdb2gmx changed a few residue names and atom names to conform to the names used in the AMBER03 .rtp file (found within ~/share/gromacs/top/amber03.ff/ of your local GROMACS directory). Ensure that the changes make sense, and it is okay to proceed. |
Revision as of 10:46, 3 August 2011
Contents
Purpose
The purpose of this tutorial is to provide an introduction to the fundamental commands needed to set up, run, and analyze MD simulations in GROMACS. This tutorial assumes you have already correctly installed GROMACS. This tutorial was written using GROMACS 4.5.4.
Download and Prepare PDB File
Ubiquitin is a small and perfectly ordinary protein, making it ideal for an introductory tutorial. A crystal structure can be found at the PDB website under the accession code 1UBQ. Before proceeding to the next step, now is a good time to use your favorite text editor to check for a few things:
1.) Are crystal waters present in the PDB file? In the case of 1UBQ, there are many crystal waters. Ubiquitin is non-enzymatic, though, so the waters are not important for an active site mechanism, for example. It is a good idea to delete the waters now and allow the GROMACS solvation tool to fill them back in later.
2.) Are there any ligands or non-standard residues present in the PDB file? In the case of 1UBQ, there are no ligands or non-standard residues. Ligand preparation and inclusion is covered in another tutorial (MD Simulation: Protein-Ligand Complex). Without going into too much detail, non-standard residues are okay so long as the residue name and atom names conform to the corresponding entry in the residue topology file (.rtp) of the force field you choose.
3.) Are there any residues with missing atoms in the PDB file? In the case of 1UBQ, there are not. If there were, however, you would need to take extra preparation steps beforehand to fix the broken residues or, if they are at one of the protein termini, consider removing them from the file.
GROMACS Coordinate and Topology Files
The first step in MD simulation with GROMACS is to create GROMACS-compatible coordinate (.gro) and topology (.top) files. Strictly speaking, .pdb files are GROMACS-compatible, but we might as well convert to a .gro coordinate file to keep in line with the spirit of the tutorial. The command line execution looks like this:
pdb2gmx -f 1UBQ.pdb -o protein.gro -p topol.top -ignh -v
When prompted, choose '1' for the AMBER03 force field, and '1' again for the TIP3P water model. These selections are fine for this tutorial, but make sure you think very carefully about your choice before picking a force field in your research. You may ask yourself, should I use an all-atom force field or a united-atom force field? Is my force field selection compatible with lipids or small molecules that I want to use? Can I compare the results of simulations using my force field to other simulations I performed previously / found in the literature? etc.
After you have picked the force field and a solvent model compatible with that force field, it is a good idea to read through the output on the screen to make sure there are no Errors or Warnings. In this case, it looks like there actually was one Warning:
WARNING: there were 0 atoms with zero occupancy and 28 atoms with
occupancy unequal to one (out of 602 atoms). Check your pdb file.
A quick look back at the original PDB file reveals that the four residues at the C-terminus of ubiquitin in 1UBQ were not as well resolved as the rest of the protein. Pdb2gmx noticed that, too, and adjusted the occupancy of each to '1'. This should be okay for now, as the atomic positions of these residues will be refined later in the minimization and equilibration steps. Aside from that one warning, it appears pdb2gmx changed a few residue names and atom names to conform to the names used in the AMBER03 .rtp file (found within ~/share/gromacs/top/amber03.ff/ of your local GROMACS directory). Ensure that the changes make sense, and it is okay to proceed.
There are three output files resulting from pdb2gmx:
1.) protein.gro # gromacs-format coordinate file 2.) topol.top # gromacs-format topology file 3.) posre.itp # position restraint file
It is very important to know and understand the contents of each file before continuing. Make sure you look through each file until you are able to make sense of the information contained within each.
Coordinate file
The GROMACS coordinate file contains the following:
Ubiquitin from 1UBQ - no water molecules (yet)
1231
1MET N 1 2.734 2.443 0.261
1MET H1 2 2.807 2.485 0.207
1MET H2 3 2.771 2.413 0.349
...
76GLY C 1229 4.003 3.999 3.543
76GLY OC1 1230 4.086 3.957 3.625
76GLY OC2 1231 3.893 4.052 3.569
3.06857 3.23738 3.66170
GROMACS coordinate files always contain three special lines. The first line is a title - it is good practice to use a detailed title specific to the system being simulated. The second line is the number of atoms. The final line contains the box vectors (in nanometers). The lines in between contain the residue number, residue name, atom name, atom number, and cartesian coordinates (in nanometers) for each atom in the system.
Topology file
The GROMACS topology file contains the following sections:
#include "amber03.ff/forcefield.itp"
[ moleculetype ] Protein_chain_A 3 [ atoms ] ... [ bonds ] ... [ pairs ] ... [ angles ] ... [ dihedrals ] ... [ dihedrals ] ... #ifdef POSRES #include "posre.itp" #endif
[ system ] Ubiquitin [ molecules ] Protein_chain_A 1
The first section is an include statement that, when this file is processed, pastes the bonded and non-bonded information specific to your force field directly into the topology file. It is a good idea to familiarize yourself with the contents of the 'forcefield.itp' file, and the files included within it, all found under ~/share/gromacs/top/amber03.ff/.
The second section (usually) contains 6 or 7 sub-sections. First, under '[ moleculetype ]' is the name of the molecule followed by a number. In this case, it is by default called 'Protein_chain_A'. The number N indicates that for this molecule, exclude non-bonded interactions of all bonded neighbors up to N bonds away. Unless you have a very good idea of what you are doing, you should not change this number. A given moleculetype is followed by (and it must be in this order) atom information, bond information, pair-wise exclusion information, angles information, dihedral information, improper dihedral information, and, optionally, position restraint information. Not all of the sections must be present. Further, this entire section is repeated for each type of molecule that you have. For example, if you are simulating a protein in water, you will have two consecutive '[ moleculetype ]' sections.
The final section only includes two sub-sections. Under '[ system ]' is a system title chosen by the user, and under '[ molecules ]' is a list of the moleculetypes found in the topology file, followed by the number of times each moleculetype appears in the coordinate file. For example if you have a coordinate file that contains a protein in 10,000 water molecules, your topology file should look roughly like:
#include "amber03.ff/forcefield.itp"
[ moleculetype ] Protein 3 ...
[ moleculetype ] SOL 1 ...
[ system ] Protein in water [ molecules ] Protein 1 SOL 10000
where the moleculetypes under '[ molecules ]' are listed in the same order as they appear in the topology file. Note that the topology file you generated already contains an #include statement to include the moleculetype for TIP3P water and for ions, but because there are no water molecules or ions in our coordinate file, they are not yet listed under '[ molecules ]'. Chapter 5 of the GROMACS manual is an excellent resource for further information regarding molecular topologies.
Position restraints file
The final file that was created by pdb2gmx is the position restraints file:
[ position_restraints ]
1 1 1000 1000 1000
5 1 1000 1000 1000
7 1 1000 1000 1000
...
Each line indicates an atom number, a functional form (1 = harmonic restraint), and force constants in cartesian space. By default, only the protein backbone atoms are listed in this file. Looking back at the topology file, you can see that if 'POSRES' is defined when you begin simulation, then these parameters will be included in the molecular topology, thus restraining the backbone of the protein during simulation. If 'POSRES' is not defined, then the position restraints are ignored.
Box Preparation
In the next few steps, you will define a box size for your system, center the protein in the box, solvate, and finally, add counterions to the system. The files you need to start this step are:
1.) protein.gro # gromacs-format coordinate file 2.) topol.top # gromacs-format topology file 3.) posre.itp # position restraint file
editconf
The GROMACS tool editconf is very useful to change the format of your coordinate files, to rotate and translate coordinate files, to define the box size, among other things. Typing 'editconf -h' will display a brief description of its capabilities. To create a rectangular box around the protein, type:
editconf -f protein.gro -o protein_box.gro -bt triclinic -d 1.2 -c
The only input needed is the coordinate file generated previously. With '-bt triclinic' you are choosing to create a rectangular box. It is important to note that rectangular boxes can be very inefficient, especially for globular proteins like ubiquitin. In this tutorial, we will keep with the rectangular box, but in the future, I would highly recommend using '-bt dodecahedron' for globular proteins. The option '-d 1.2' creates a buffer of 1.2 nm between the outside of the protein and the edge of the box, and '-c' centers the protein in the box and puts the corner of the box at {0, 0, 0} in cartesian space. The output file is called 'protein_box.gro'.
The most important question to ask now is why choose 1.2 nm? The short answer is that you don't want the protein to 'see' its periodic image across the boundary of the box. If you refer to the original paper for the AMBER 03 force field (Duan, et al., J. Comput. Chem. 2003, 24, 1999-2012), they use short-range VDW and electrostatic cut-offs of 0.8 nm - when you begin simulating, it is very important to do the same. In order for the protein to avoid seeing its image across the periodic boundary, it must be at least twice the cut-off distance from the next nearest image of itself. That being said, the space between the protein and the edge of the box only really needs to be 0.8 nm, but it is still a good idea to include extra space especially if there is a chance the protein might unfold a little bit or if is an oblong shape. It is better to have a slightly larger box size now than to find out later that your protein was interacting with its periodic image during the simulation. (See the image below).
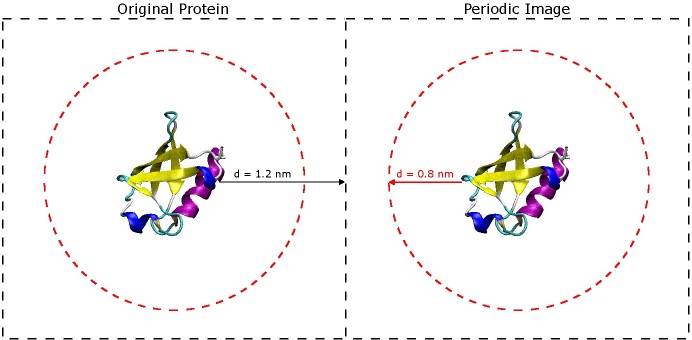 The dashed black squares indicate the box boundaries, and the dashed red circles indicate the short-range VDW and electrostatic cut-offs. So long as the red circles do not overlap, the protein will not be able to "see" its periodic image.
The dashed black squares indicate the box boundaries, and the dashed red circles indicate the short-range VDW and electrostatic cut-offs. So long as the red circles do not overlap, the protein will not be able to "see" its periodic image.
genbox
The next GROMACS command, 'genbox', will fill the box with solvent molecules.
genbox -cp protein_box.gro -cs spc216.gro -o protein_sol.gro -p topol.top
The '-cp' flag specifies the coordinate file of the system to be solvated, and '-cs spc216.gro' tells genbox to use an SPC (a simple three-point water model) coordinate file to populate the box. Thee output coordinate file is called 'protein_sol.gro'. The topology file, 'topol.top', is also provided on the command line. If you check the end of the topology file, you can see that it has been updated in the following way:
[ molecules ] Protein_chain_A 1 SOL 5760
At this point, it is useful to open 'protein_sol.gro' in VMD to ensure that the protein is correctly placed in the center of the box and there are no strange solvent artifacts.
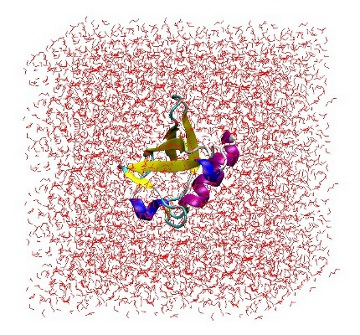 1UBQ in a triclinic box solvated with 5,760 water molecules.
1UBQ in a triclinic box solvated with 5,760 water molecules.
genion
The final step before simulation is to add enough ions to the system to neutralize the net charge or, alternatively, add enough ions to neutralize the net charge and reach some physiological concentration. If you are simulating a system in an attempt to replicate some experimental observable, for example, it is important to use the same salt concentration in your system as is used in the experiment. A standard value for salt concentration often used to replicate human physiology is 100 mM.
The 'genion' tool searches through your coordinate file and will randomly replace water molecules with ions. In order for it to work, however, it requires a pre-processed input file (with extension .tpr) that contains all of the information from both the coordinate and topology files. In order to generate such a file, you will use the tool 'grompp'. Aside from the coordinate and topology file, 'grompp' also requires that you provide a MD parameter file (.mdp) as input. Because the .tpr file you are generating is not going to be used for dynamics, rather it is just going to be used to add ions, the contents of the .mdp file are not important. Nevertheless, it still must be provided on the command line. An example .mdp file with all of the default parameters can be found here: genion.mdp. To create the input file, execute:
grompp -f genion.mdp -c protein_sol.gro -p topol.top -o genion_input.tpr
As always, read what was output to the screen, and if there are no major Errors or Warnings, than it is okay to proceed. The net charge of the ubiquitin system is already 0 (a running total of the charge can be found in the '[ atoms ]' section of the protein moleculetype under the definition 'qtot'), so instead of neutralizing the system, add enough NaCl to reach 100 mM salt concentration. To do so, execute:
genion -s genion_input.tpr -p topol.top -o protein_sol_nacl.gro -pname NA -pq 1 -nname CL -nq -1 -conc 0.1 -neutral
It is a good idea to read the output from 'genion -h' in order to gain a full understanding of the command line options. When prompted, choose group 13 (SOL). Genion then chooses 22 solvent molecules and replaces half with sodium ions, and the other half with chloride ions. The topology file is also provided on the command line so that it may be updated accordingly. The '[ molecules ]' section has been modified by removing 22 SOL molecules and adding 11 each of NA and CL molecules.
[ molecules ] Protein_chain_A 1 SOL 5738 NA 11 CL 11
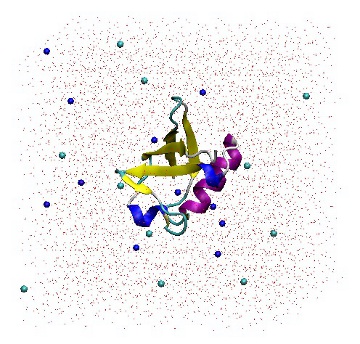 1UBQ in a triclinic box solvated with 5,738 water molecules and 22 ions.
1UBQ in a triclinic box solvated with 5,738 water molecules and 22 ions.
Energy Minimization
Prior to running actual dynamics, you will need to perform an energy minimization. The purpose of a minimization is to relax the molecular geometry of the system in order to get rid of any atomic clashes or other irregularities that may exist. The files you need to start this step are:
1.) protein_sol_nacl.gro # gromacs-format coordinate file 2.) topol.top # gromacs-format topology file 3.) posre.itp # position restraint file 4.) ubq_min.mdp # energy minimization parameter file
The file 'ubq_min.mdp' contains the run parameters for the minimization. A copy of this file can be found here: ubq_min.mdp. The comments in the file help to explain the purpose of each parameter. However, a review of Chapter 7 in the GROMACS manual would be invaluable (specific sections are listed in the .mdp file). In brief, this parameter file calls for a steepest descent energy minimization not to exceed 2,000 steps. The minimization will be considered successful if the maximum force on any atom is less than 1000 kJ/mol/nm, at which point the minimization will stop. Other parameters of note include periodic boundary conditions in the x-, y-, and z-directions, 0.8 nm cut-offs for short-range VDW and electrostatic interactions, and particle-mesh Ewald long-range electrostatics. If any of these settings do not make sense, it is unwise to proceed without reading about them in Chapter 7 of the manual.
The main integration engine of GROMACS is a tool called 'mdrun'. As input, it requires a pre-processed run input file (.tpr) that contains the system's coordinates, topology, and parameters for the minimization itself. As before, 'grompp' is used to generate that file. Execute the following command:
grompp -f ubq_min.mdp -c protein_sol_nacl.gro -p topol.top -o input_min.tpr
Read the screen output carefully. If the .tpr file is generated successfully, you can perform the minimization:
mdrun -s input_min.tpr -deffnm ubiquitin_min -v
By using the '-v' option, you can watch the minimization progress on the screen. Using the steepest descent method, small systems may equilibrate after only a few hundred steps; larger systems may may take several thousand steps. It mostly depends on the complexity of the system and the quality of the original starting structure. Once the minimization is complete (which should take around 500 steps), you will find that several files have been written to disk, all with the prefix 'ubiquitin_min'. They are as follows:
1.) ubiquitin_min.gro # system coordinates at the final step 2.) ubiquitin_min.trr # trajectory file 3.) ubiquitin_min.log # log file 4.) ubiquitin_min.edr # energy file
The first file, 'ubiquitin_min.gro', contains the fully minimized coordinates of the system (presuming the minimization was successful). The second file, 'ubiquitin_min.trr' contains a trajectory of the minimization. The frequency with which frames are written to the trajectory is specified in the .mdp file that was passed to grompp. You can view this in VMD by first loading the original coordinate file (protein_sol_nacl.gro), followed by the trajectory file. The log file contains information about the run parameters used for the minimization, as well as various system energies during the minimization. It is convenient to check the progress of long MD simulations by following the end of this file. Finally, ubiquitin_min.edr is a binary energy file that can be read by the GROMACS tool 'g_energy', yielding many pieces of valuable information. In this case, you can use it to check the potential energy of the system as a function of minimization step. Execute the following command:
g_energy -f ubiquitin_min.edr -o energy.xvg
A prompt will appear asking for a specific term you would like to analyze:
Select the terms you want from the following list by selecting either (part of) the name or the number or a combination. End your selection with an empty line or a zero. ------------------------------------------------------------------- 1 Bond 2 Angle 3 Proper-Dih. 4 Improper-Dih. 5 LJ-14 6 Coulomb-14 7 LJ-(SR) 8 Disper.-corr. 9 Coulomb-(SR) 10 Coul.-recip. 11 Potential 12 Pres.-DC 13 Pressure 14 Vir-XX 15 Vir-XY 16 Vir-XZ 17 Vir-YX 18 Vir-YY 19 Vir-YZ 20 Vir-ZX 21 Vir-ZY 22 Vir-ZZ 23 Pres-XX 24 Pres-XY 25 Pres-XZ 26 Pres-YX 27 Pres-YY 28 Pres-YZ 29 Pres-ZX 30 Pres-ZY 31 Pres-ZZ 32 #Surf*SurfTen 33 Mu-X 34 Mu-Y 35 Mu-Z 36 T-rest
Choose '11' for the potential energy, and hit return on an empty line to finish. A file called 'energy.xvg' will be written to file. This is a simple two-column data file that can be plotted in a program such as Grace:
 Potential energy of system as a function of minimization step.
Potential energy of system as a function of minimization step.
Convergence of the potential energy in a system to a large negative number (on the order of ~10^5) is a very good indication that the energy minimization was successful. If you find that for your system the potential energy has converged, and if the maximum force on any atom is below a reasonable tolerance cut-off, then it is okay to proceed to Equilibration MD. Otherwise, you may need to use other minimization methods (such as conjugate gradients or L-BFGS) to reach a lower energy well. The implementation of these are described in detail in the GROMACS manual.
Equilibration MD
Equilibration will be carried out in two steps. First, an NVT (constant Number of atoms, Volume, and Temperature) simulation will be performed in order to bring the system to the target temperature. Second, an NPT (constant Number of atoms, Pressure, and Temperature) simulation will be performed to allow the system to find the correct density.
NVT Simulation
A parameter file for an NVT simulation can be found here: ubq_nvt.mdp. This file calls for an MD run of 25,000 steps with a 2 fs timestep (a total of 50 ps). The 'define = -DPOSRES' line initiates the backbone restraints on the protein, which should remain on until production MD. Temperature coupling is performed using the Berendsen method. In order to be sure velocities (and therefore temperature) are evenly distributed across all of the molecule types, it is a good idea to couple protein atoms and non-protein atoms to separate baths. Without separate coupling, you run the risk weird phenomena such as a system where the water molecules are at 350 K and the protein is at 200 K. Finally, all bonds are constrained using the linear constraint solver (LINCS) algorithm. Review the GROMACS manual if you are unfamiliar with the implementation or purpose of any of these parameters.
Execute the command:
grompp -f ubq_nvt.mdp -c ubiquitin_min.gro -p topol.top -o input_nvt.tpr
Read the output carefully. If it is okay to proceed, then:
mdrun -s input_nvt.tpr -deffnm ubiquitin_nvt -v
This step will take longer than the energy minimization. However, GROMACS automatically threads an 'mdrun' job across all of the processors available on the host machine. If you executed this command on George, it will try to use all 8 processors and the job should only take about 5-10 minutes. If you would like to run 'mdrun' in parallel on cluster, the command line should look something like:
mpirun -n 16 mdrun -s input_nvt.tpr -deffnm ubiquitin_nvt -np 16
A good rule of thumb is that for a given system you will achieve the most efficient scaling if you use N processors, where N = <# of atoms> / 1000. At the completion of the job, you will find the 4 types of output files that you saw following energy minimization, in addition to 2 new output files:
1.) ubiquitin_nvt.gro # system coordinates at the final step 2.) ubiquitin_nvt.trr # full-precision trajectory file 3.) ubiquitin_nvt.log # log file 4.) ubiquitin_nvt.edr # energy file 5.) ubiquitin_nvt.xtc # compressed trajectory file 6.) ubiquitin_nvt.cpt # checkpoint file
The .xtc file is another trajectory file. For the same number of frames, the .xtc file is much smaller than the .trr file because it contains only atomic coordinates, whereas the .trr file also contains full-precision velocities and forces, which are only useful for continuing runs. In practice, you may find it most convenient to only write to the .trr file once at the end of a simulation, and to write to the .xtc file much more frequently (e.g., every 1 ps) for analysis. To view the trajectory, load the original coordinate file (ubiquitin_min.gro) into VMD, followed by the .xtc file. The final file, 'ubiquitin_nvt.cpt', is a new-ish implementation in GROMACS that renders the .trr file obsolete. The checkpoint file is by default written to disc every 15 minutes (and always at the very end of a simulation), and it contains the latest full-precision velocities, forces, and energies that are required to continue a simulation, or restart a crashed-simulation. This will be demonstrated in the next section, but first, it is important to verify that the ubiquitin system equilibrated around the target temperature. Use 'g_energy' again, and this time choose '14' for Temperature.
g_energy -f ubiquitin_nvt.edr -o energy.xvg
Visualize the resulting plot in Grace:
 System temperature as a function of time during NVT equilibration.
System temperature as a function of time during NVT equilibration.
From this plot it is apparent that the system is well equilibrated around the target temperature (310 K), and it is okay to proceed to the NPT equilibration step.
NPT Simulation
Following equilibration to a target temperature, you must next relax your system into a constant pressure ensemble. A parameter file for NPT simulation can be found here: ubq_npt.mdp. As you can see, this parameter file calls for a run of 250,000 steps with a 2 fs timestep (a total of 500 ps). Position restraints are still being applied to the backbone. The Berendsen thermostat has been replaced with a Nose-Hoover thermostat, which will produce a more correct ensemble of kinetic energies. The Nose-Hoover thermostat, however, must be implemented only after the system is already near its target temperature (which was achieved by the Berendsen thermostat), or the kinetic energy of the system will fluctuate wildly. Because this run will be a continuation of the previous run, 'gen_vel' has been set to 'no'. Finally, the Parrinello-Rahman method is used to couple pressure isotropically (the same in all directions) to a value of 1.0 bar. As soon as you have a thorough understanding of the logic behind each setting in the parameter file, pre-process your run input file with grompp:
grompp -f ubq_npt.mdp -c ubiquitin_nvt.gro -t ubiquitin_nvt.cpt -p topol.top -o input_npt.tpr
The important difference with this invocation of 'grompp' is that you specified '-t ubiquitin_nvt.cpt'. This flag indicates that the forthcoming run will be a continuation of the previous run, taking into account the full-precision velocities and forces on all atoms from the checkpoint file. If there are no Errors, then:
mdrun -s input_npt.tpr -deffnm ubiquitin_npt -v
This run requires many more steps than the NVT equilibration did, and it may take an hour or so if you run it on George. When complete. you will have the same 6 types of output files as those that resulted from the NVT simulation. Use g_energy again to tell whether the goal of the equilibration was acheived:
g_energy -f ubiquitin_npt.edr -o energy.xvg
Choose '22' for density and visualize the resulting plot in Grace:
 System density as a function of time during NPT equilibration.
System density as a function of time during NPT equilibration.
The system appears to have quickly equilibrated very near to a physiological value. By adding the following flags to g_energy:
g_energy -f ubiquitin_npt.edr -b 250
and again choosing '22', the program reports a density of 1002.1 kg/m^3 as averaged over the last 250 ps of simulation. At this point, it is okay to move on to production MD.
Production MD
The parameter file for production MD looks almost exactly like that of the NPT equilibration step with one important exception: position restraints on the backbone have been lifted. The parameter file can be found here: ubq_md.mdp. This file calls for 2,500,000 MD steps with a 2 fs timestep, totaling 5,000 ps (or 5 ns) of sampling. Other than the longer sampling time and the release of restraints, there are no other differences between this simulation and the NPT equilibration. To pre-process the run input file, execute:
grompp -f ubq_md.mdp -c ubiquitin_npt.gro -t ubiquitin_npt.cpt -p topol.top -o input_md.tpr
Remember, it is important to provide the checkpoint file on the command line as this is a continuation of the previous run. If there are no Errors, then begin simulation with:
mdrun -s input_md.tpr -deffnm ubiquitin_md
Running this simulation on George may take 10-12 hours. It will take significantly less time if you run it in parallel on cluster. Examine the last 200 or so lines of the .log file to find some statistics on the speed of the run, as well as a very detailed decomposition of where all of the computation time was spent:
R E A L C Y C L E A N D T I M E A C C O U N T I N G
Computing: Nodes Number G-Cycles Seconds %
-----------------------------------------------------------------------
Domain decomp. 8 2500000 91520.964 32768.2 11.3
DD comm. load 8 2499011 699.920 250.6 0.1
DD comm. bounds 8 2499001 1137.751 407.4 0.1
Comm. coord. 8 2500001 3508.266 1256.1 0.4
Neighbor search 8 2500001 232396.884 83207.4 28.8
Force 8 2500001 204567.718 73243.5 25.3
Wait + Comm. F 8 2500001 4545.445 1627.5 0.6
PME mesh 8 2500001 213796.271 76547.7 26.5
Write traj. 8 5041 74.665 26.7 0.0
Update 8 2500001 14321.922 5127.8 1.8
Constraints 8 2500001 27057.311 9687.6 3.3
Comm. energies 8 2500002 3582.277 1282.6 0.4
Rest 8 11075.937 3965.6 1.4
-----------------------------------------------------------------------
Total 8 808285.331 289398.7 100.0
-----------------------------------------------------------------------
-----------------------------------------------------------------------
PME redist. X/F 8 5000002 45742.470 16377.6 5.7
PME spread/gather 8 5000002 94963.334 34000.7 11.7
PME 3D-FFT 8 5000002 52335.780 18738.3 6.5
PME solve 8 2500001 20644.201 7391.5 2.6
-----------------------------------------------------------------------
Parallel run - timing based on wallclock.
NODE (s) Real (s) (%)
Time: 36174.836 36174.836 100.0
10h02:54
(Mnbf/s) (GFlops) (ns/day) (hour/ns)
Performance: 133.592 11.329 11.942 2.010
Finished mdrun on node 0 Fri Jul 22 11:24:04 2011
Extending a run requires two simple steps. First, the run input file is modified to specify a longer simulation time. Second, 'mdrun' is called again with some additional flags. For example, if you would like to extend this run by 5,000 ps, you would execute the following:
tpbconv -s input_md.tpr -o input_md_extended.tpr -extend 5000 mdrun -s input_md_extended.tpr -deffnm ubiquitin_md -cpi ubiquitin_md.cpt -append
The first command uses the tool 'tpbconv' to add 5,000 ps of simulation time to the .tpr file. Of course the output file name (specified with the '-o' flag) is arbitrary. In fact, the output file name can be the same as the input file name. GROMACS would automatically backup the original file and you would not need to worry about keeping track of multiple .tpr files. The '-append' flag on the second line tells 'mdrun' to append all new calculations to the end of the original output files from the first 5 ns simulation. In fact, if your run were to die because the server crashes, for example, you would just need to execute the 'mdrun' command above with your original .tpr file in order to restart it.
Analysis
GROMACS comes equipped with many analysis tools, a complete list of which can be found in the manual. Here you will be exposed to a few useful analysis tools: 'g_rms', 'g_rmsf', and 'do_dssp'. But first, it is useful to learn how to process your trajectory file to only keep the components you are interested in. For example, start with the two files:
1.) input_md.tpr # run input file for production MD 2.) ubiquitin_md.xtc # full system 5 ns trajectory
The next three analyses that you are going to perform only require protein coordinates, not water / ion coordinates. Use 'trjconv' to remove water molecules and ions from the .xtc file, then use 'tpbconv' to remove water / ion information from the .tpr file:
trjconv -s input_md.tpr -f ubiquitin_md.xtc -o protein.xtc tpbconv -s input_md.tpr -o protein.tpr
In each case, you will be prompted to choose a group. Choose group '1 (Protein)' both times. Now, use 'trjconv' again to dump out the first frame of the trajectory:
trjconv -s protein.tpr -f protein.xtc -o protein.gro -dump 0
Choose either '0 (System)' or '1 (Protein)' to dump all protein coordinates (with these modified input files, the protein is the whole system). The '-dump 0' command will write out the coordinates closest to 0 ps. At this point, it is a good idea to open protein.gro in VMD, followed by protein.xtc in order to watch the trajectory and make sure nothing funny happened during the simulation. In the production MD parameter file, you turned on center-of-mass motion removal. That is why the protein tends to stay in the center of the box during simulation. However, you will notice that it still rotates around all three axes freely. To remove that, you simply need to perform a best-fit translation and rotation calculation on each frame of the trajectory using the starting coordinates as the reference structure. To do so, execute:
trjconv -s protein.tpr -f protein.xtc -o protein_fit.xtc -fit rot+trans
It will prompt you for a group to use when calculating the least-squares fit. Traditionally, the backbone is used for such a calculation, so choose '4 (Backbone)'. Then it will ask which group you would like to write to disk. Choose '0 (System)' or '1 (Protein)'. Load this modified trajectory into VMD to see the effect.
RMSD
Root mean square deviation (RMSD) is a measure of how much the protein structure changes over the course of the simulation. Changes on the order of 1-3 Angstroms are perfectly acceptable (and expected) for small, globular proteins. Changes much larger than that, however, indicate that your protein may be misbehaving during the simulation. The other thing to look for is convergence. If the RMSD of your protein is still changing at the end of your simulation (trending upwards, usually), then your simulation is probably not long enough. To measure RMSD, use the GROMACS tool 'g_rms':
g_rms -s protein.tpr -f protein_fit.xtc -o rmsd.xvg
Choose '4 (Backbone)' for the least squares fit, and '4 (Backbone)' again for the RMSD measurement. The reference structure is taken from 'protein.tpr'. Each frame in protein_fit.xtc is iteratively fit to the reference structure (because you already did this in the previous step, you can turn the fitting off by specifying '-fit none' on the command line). The output file will be called 'rmsd.xvg', which can be visualized in Grace.
 Ubiquitin backbone RMSD during a 5 ns NPT production simulation.
Ubiquitin backbone RMSD during a 5 ns NPT production simulation.
The black line represent the RMSD of the protein backbone during the simulation, which remains within an acceptable range. A look at the running average of the RMSD (red line) indicates that, despite this being only a 5 ns simulation, the RMSD is reasonably stable around 0.11 nm.
RMSF
RMSD is a measure of global backbone deviation. If you are interested in more local changes, you can measure the root mean square fluctuation (RMSF). The GROMACS tool 'g_rmsf' was written for that purpose. Execute the following command:
g_rmsf -s protein.tpr -f protein_fit.xtc -o rmsf.xvg -oq rmsf.pdb -res
Now you must make a decision. Are you more interested in backbone fluctuation, sidechain fluctuation, entire residue fluctuation, or something completely different? What you pick here will probably change from project to project depending on the scientific question you are trying to ask. In the case of ubiquitin, you may ask yourself, why were the occupancy values of the last four residues less than 1? (If you remember back to the crystal structure and 'pdb2gmx', you were presented with a Warning about this exactly). The occupancy values in the crystal structure applied to entire residues - not just the backbone or just the sidechain. Therefore, choose '1 (Protein)', in order to measure fluctuations of whole residues (the '-res' flag will group the output by whole residues rather than by individual atoms). There are two important output files:
1.) rmsf.xvg # a data file of RMSF values plotted by residue number that can be viewed in Grace 2.) rmsf.pdb # a structure file that contains beta-factors equal to RMSF values
The first file should be viewed in Grace:
 Ubiquitin backbone RMSF averaged over 5 ns simulation.
Ubiquitin backbone RMSF averaged over 5 ns simulation.
On this plot, peaks indicate areas of the protein that fluctuated the most during the simulation. The C-terminal tail fluctuated much more than any other part of the protein, helping to explain why they crystallographers assigned such a low resolution to that area.
Open the second file, 'rmsf.pdb', in VMD and color residues by Beta. The result should look like the following:
 Ubiquitin structure colored by RMSF. Red values indicate areas of more fluctuation, blue values indicate areas of less fluctuation.
Ubiquitin structure colored by RMSF. Red values indicate areas of more fluctuation, blue values indicate areas of less fluctuation.
Now it is easy to tell that the C-terminus of the protein fluctuates the most, and it appears that the alpha-helix is the most stable part of the protein.
As a final note on RMSF analysis, you must be aware that the RMSF plot will change depending on what part of the simulation you choose to analyze. For example, the plot seen above is the average fluctuation of each residue over the entire simulation. In practice, that is not the best way to measure RMSF. At the beginning of the simulation, certain residues may be fluctuating more as they transition from the crystal form of the protein to a fully solvated and dynamic form. It is better to simulate for several nanoseconds, and when the RMSD converges, use that as a starting point for RMSF measurements. For example if you want to measure RMSF beginning from 4 ns, the command would be:
g_rmsf -s protein.tpr -f protein_fit.xtc -o rmsf.xvg -oq rmsf.pdb -res -b 4000
DSSP
DSSP stands for Dictionary of Secondary Structure of Proteins (or something similar). The purpose of this analysis is to measure the secondary structure content of a protein as a function of time. The analysis breaks down secondary structure assignments (i.e. helix, sheet, turn, etc.) by individual residues for each time step, allowing you to visualize or quantify the data in meaningful ways. The GROMACS tool 'do_dssp' uses the DSSP library and executable from this website to measure secondary structure in your trajectory file. George already has the appropriate DSSP library and executable installed. All you have to do to use dssp is issue 'do_dssp', and GROMACS will automatically find the library and executable. Type the following:
do_dssp -f protein.xtc -s protein.tpr -sc scount.xvg -o ss.xpm -dt 10
When prompted, you must choose '5 (Mainchain)'. The trajectory saved snapshots at every 1 ps, which is a little too frequent for this type of analysis, so use the option '-dt 10' to only look at every 10th frame. The first output file, scount.xvg, can be visualized in Grace by typing:
xmgrace -nxy scount.xvg
 Ubiquitin secondary structure content
Ubiquitin secondary structure content
As an added bonus, the end of the file 'scount.xvg' contains the following two lines:
# Totals 26169 9107 11705 1009 1506 7064 6391 1294 # SS % 0.69 0.24 0.31 0.03 0.04 0.19 0.17 0.03
where:
@ s0 legend "Structure" @ s1 legend "Coil" @ s2 legend "B-Sheet" @ s3 legend "B-Bridge" @ s4 legend "Bend" @ s5 legend "Turn" @ s6 legend "A-Helix" @ s7 legend "3-Helix"
This is a very convenient way to analyze average secondary structure content of a protein. This could be especially useful when trying to predict the secondary structure of unstructured proteins as well.
The second file, ss.xpm, contains a breakdown of secondary structure for each residue as a function of time step. Although some programs can open this type of file, it is generally easier to process it with the GROMACS tool 'xpm2ps' first. Make a copy of this file: ps.m2p, then execute the following:
xpm2ps -f ss.xpm -di ps.m2p -o ubq_ss.eps
The tool 'xpm2ps' has quite a lot of options. You can type 'xpm2ps -h' or check the GROMACS website to see how else you can render postscript images from xpm data. The version of ps.m2p that you downloaded will render the following image:
 Ubiquitin secondary structure content by residue
Ubiquitin secondary structure content by residue
From this plot it is easy to see the stability (or de-stability) of secondary structure elements as a function of time. For example, it appears a stretch of residues from about 55-60 mostly are in a 'turn' conformation, but sporadically adopt an 'alpha-helix' or '3-helix' conformation as well.