2017 Denovo refine tutorial with PDB 1BJU
Contents
2017 De novo refinement tutorial with PDB 1BJU
The De novo Refinement module of DOCK constructs new ligand molecules by adding side chains to any atom of a reference molecule. The side chains are taken from a library of fragments that will be specified as an input by the user. The atom at which the side chain will be added needs to be specified as a 'dummy' atom. This is done by opening the pdb file and modifying the atom type to "Du" (column 6).
Make sure to complete the 2016 Dock tutorial with Beta Trypsin. Some of the output files generated from that tutorial will be required below.
In the previous dock tutorial directory, create a directory for the de novo refinement run named "04.denovo_refinment".
The Files Needed For De novo Refinement
To run the de novo refinement code with single grid scoring you need these files:
From dock tutorial:
03.box-grid/grid 01.dockprep/1BJU.lig.mol2
New files:
PATH/gen-frags-12/fraglib_scaffold.mol2 PATH/gen-frags-12/fraglib_linker.mol2 PATH/gen-frags-12/fraglib_sidechain.mol2 PATH/gen-frags-12/fraglib_torenv.dat PATH/dock6/parameters/vdw_AMBER_parm99.defn PATH/dock6/parameters/flex.defn PATH/dock6/parameters/flex_drive.tbl
The first four files are in a fragment library directory called "gen-frags-12" which can be generated by the user here: Fragment Library Generation. The bottom three files are typical parameter files contained in the parameters directory in DOCK6.
Preparing The Files
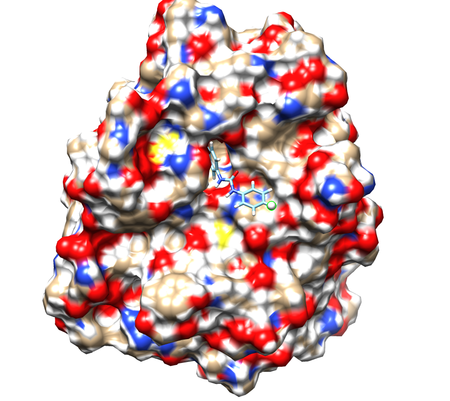
Start a Chimera session and click Open and Select the 1BJU.rec.mol2 to open the receptor file. Show surface by clicking Actions->Surface->show. Display charge distribution on surface by Actions->Color->by heteroatom. Open ligand file 1BJU.lig.mol2. To select an atom to act as a dummy for side chains attachment, find an atom in a position that allows addition of molecular groups, in other words, not inside binding pocket and not tightly packed against receptor atoms. Here, we select H12 to be our dummy atom (see figure).
Copy the ligand file 1BJU.lig.mol2 into 1BJU.lig.dummyH.mol2. Open 1BJU.lig.dummyH.mol2 with vi and modify the type of atom 28 H12 to Du (in column 6). Make sure the spacing after the modification stays aligned with the other columns. Save file.
De novo Refinement Run
Make input file by typing: touch dnref.in Copy the following commands into this file. Alternatively, one can create this file interactively by calling dock with the empty dnref.in file. Dock will ask for each input specification one by one.
Make sure to include the correct paths to the files for the fragment library and the Dock parameter files.
conformer_search_type nova
dn_fraglib_scaffold_file {PATH_TO_FILE}/gen-frags-12/fraglib_scaffold.mol2
dn_fraglib_linker_file {PATH_TO_FILE}/gen-frags-12/fraglib_linker.mol2
dn_fraglib_sidechain_file {PATH_TO_FILE}/gen-frags-12/fraglib_sidechain.mol2
dn_user_specified_anchor yes
dn_fraglib_anchor_file ../01.dockprep/1BJU.lig.dummyH.mol2
dn_use_torenv_table yes
dn_torenv_table {PATH_TO_FILE}/gen-frags-12/fraglib_torenv.dat
dn_sampling_method graph
dn_graph_max_picks 30
dn_graph_breadth 3
dn_graph_depth 2
dn_graph_temperature 100.0
dn_pruning_conformer_score_cutoff 100.0
dn_pruning_conformer_score_scaling_factor 1.0
dn_pruning_clustering_cutoff 100.0
dn_constraint_mol_wt 750
dn_constraint_rot_bon 15
dn_constraint_formal_charge 2.0
dn_heur_unmatched_num 1
dn_heur_matched_rmsd 2.0
dn_unique_anchors 1
dn_max_grow_layers 1
dn_max_root_size 25
dn_max_layer_size 25
dn_max_current_aps 5
dn_max_scaffolds_per_layer 1
dn_write_checkpoints yes
dn_write_prune_dump yes
dn_write_orients no
dn_write_growth_trees no
dn_output_prefix 1BJU.lig.dummyH.dnref
use_internal_energy yes
internal_energy_rep_exp 12
internal_energy_cutoff 100.0
use_database_filter no
orient_ligand no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary no
grid_score_secondary no
multigrid_score_primary no
multigrid_score_secondary no
dock3.5_score_primary no
dock3.5_score_secondary no
continuous_score_primary no
continuous_score_secondary no
footprint_similarity_score_primary no
footprint_similarity_score_secondary no
ph4_score_primary no
ph4_score_secondary no
descriptor_score_primary yes
descriptor_score_secondary no
descriptor_use_grid_score yes
descriptor_use_pharmacophore_score no
descriptor_use_tanimoto no
descriptor_use_hungarian no
descriptor_grid_score_rep_rad_scale 1
descriptor_grid_score_vdw_scale 1
descriptor_grid_score_es_scale 1
descriptor_grid_score_grid_prefix ../03.box-grid/grid
descriptor_weight_grid_score 1
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_anchor_max_iterations 500
simplex_grow_max_iterations 500
simplex_grow_tors_premin_iterations 0
simplex_random_seed 0
simplex_restraint_min no
atom_model all
vdw_defn_file {PATH_TO_FILE}/vdw_AMBER_parm99.defn
flex_defn_file {PATH_TO_FILE}/flex.defn
flex_drive_file {PATH_TO_FILE}/flex_drive.tbl
This calculation should be done quickly, and upon finishing you will have three output files:
1BJU.lig.dummyH.dnref.denovo_build.mol2 1BJU.lig.dummyH.dnref.anchor_1.prune_dump_layer_1.mol2 1BJU.lig.dummyH.dnref.anchor_1.root_layer_1.mol2
The first file contains the resulting 14 molecules. The last two files are empty since we have added only one layer (those files would be used as restart files for subsequent additions in multilayer runs).
Open Chimera to view results, use ViewDock from Tools/Surface/Binding-Analysis to check resulting molecules and rank them.
Note: If one desires to add more layers, modify the parameter of the command "dn_max_grow_layers" in the "dnref.in" input file.