2023 Denovo tutorial 1 with PDBID 4S0V
Contents
Introduction
This next section will walk you through the three options for DeNovo Design:
- Generic DeNovo Design
- Focused Fragment Design
- DeNovo Refinement
and will be a continuation of the Virtual Screening tutorial. We will continue this work with #4s0v from the Protein Data Base.
Setting Up Your Environment
For this section we will need to create some more directories following this structure:
DeNovo Refinement
The DeNovo Refinement calculation/algorithm within DOCK is an interesting way to determine the effects on a ligand/protein interaction by changing part of the small molecule. For this tutorial we are going to change just a terminal ring on our ligand and see what different residues DOCK suggests we look into further.
The overall idea behind this refinement is to delete a portion of your ligand, replace the "stub" atom (or "anchor" atom) which is now not bonded to anything with a dummy atom, and ask DOCK to find an alternative.
- Step 1: Open the ligand minimized mol2 file we generated in the previous tutorial into Chimera.
- Step 2: Open the protein into the same session
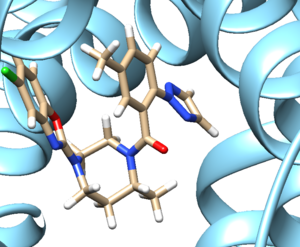
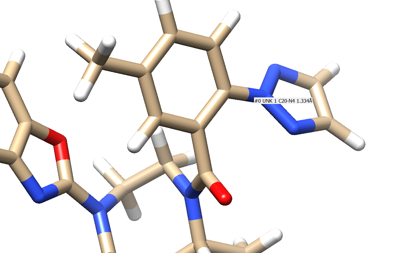
- Step 3: Examine the binding site and choose a residue on the ligand that's pointing towards the inside of the binding site. For our protein this detailed section looks like:
We see an imidazole ring pointing towards the binding site so will choose to work with that. Select the protein and hide it from view. Place your mouse over the atom connecting the ring to the rest of the ligand and note the atom and number. In this case it's N4.