2023 Denovo tutorial 1 with PDBID 4S0V
Contents
Introduction
This next section will walk you through the three options for DeNovo Design:
- Generic DeNovo Design
- Focused Fragment Design
- DeNovo Refinement
and will be a continuation of the Virtual Screening tutorial. We will continue this work with #4s0v from the Protein Data Base.
Setting Up Your Environment
For this section we will need to create some more directories following this structure:
DeNovo Refinement
The DeNovo Refinement calculation/algorithm within DOCK is an interesting way to determine the effects on a ligand/protein interaction by changing part of the small molecule. For this tutorial we are going to change just a terminal ring on our ligand and see what different residues DOCK suggests we look into further.
The overall idea behind this refinement is to delete a portion of your ligand, replace the "stub" atom (or "anchor" atom) which is now not bonded to anything with a dummy atom, and ask DOCK to find an alternative.
- Step 1: Open the ligand minimized mol2 file we generated in the previous tutorial into Chimera.
- Step 2: Open the protein into the same session
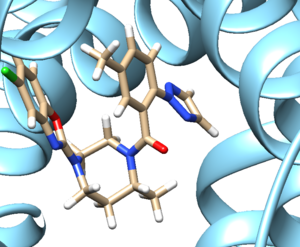
- Step 3: Examine the binding site and choose a residue on the ligand that's pointing towards the inside of the binding site. For our protein this detailed section looks like:
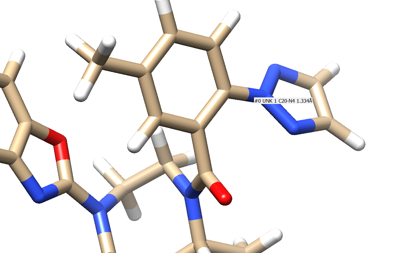
We see an imidazole ring pointing towards the binding site so will choose to work with that. Select the protein and hide it from view.
- Step 4: Place your mouse over the atom connecting the ring to the rest of the ligand and note the atom and number. In this case it's N4.
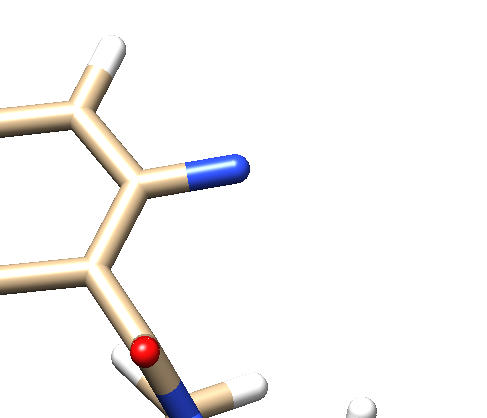
- Step 5: Delete all the atoms from N4 to the end. Your ligand should now look something like: