Pose Reproduction SB2024 V1 DOCK6.10 A
!!!!!!Under Construction!!!!!!
The purpose of this tutorial is to develop a uniform method to test pose reproduction across the Rizzo lab with the DOCK software. Note any data in this tutorial is solely for the purpose of example.
Contents
I.Introduction
-Pose reproduction is an experiment which tests a docking programs ability to predict the bound pose of a ligand to a receptor (typically a protein). An experimental structure of a protein-ligand complex is converted into 2 separate files, 1 for ligand and 1 for receptor. The docking program then predicts the binding orientation that is most energetically favorable. In the case of DOCK6, the ligand is flexibly docked with the Anchor & Grow algorithm to a rigid receptor.
-The RMSD between the docked poses and experimental pose are measured. We consider RMSD < 2 angstroms an accurate prediction. There are 3 outcomes we classify.
1) Success - The best scoring pose is < 2 angstroms
2) Scoring Fail - A pose < 2 angstroms was sampled but did not score best
3) Sampling Fail - No pose < 2 angstroms was sampled
II.Necessary files
Scripts to run Pose Reproduction in batch mode are found at:
https://github.com/rizzolab/Benchmarking_and_Validation
This tutorial uses Single Grid Energy as the primary score for docking. This is the typical score used by the Rizzo Lab for this purpose and for generating poses in Virtual Screening. Thus, grid files are required. The receptor mol2 is only necessary for visualization purposes. The list of necessary files are:
${pdb_id}.lig.am1bcc.mol2
${pdb_id}.rec.clust.close.sph
${pdb_id}.rec.clean.mol2
${pdb_id}.rec.bmp
${pdb_id}.rec.nrg
All necessary files for different versions of our test set are available for download at:
https://ringo.ams.stonybrook.edu/index.php/Rizzo_Lab_Downloads
To (re)create a testset using Rizzo Lab Protocols:
https://ringo.ams.stonybrook.edu/index.php/Test_Set_Tutorial_V1
III.Docking molecules
In 001.submit_dock.sh edit the following variables:
system_file="List of PDB codes in a file delimited by line"
testset="Path to necessary files for docking (section above)"
dock_dir="Uppermost directory of DOCK6 executable"
Additionally other variables can be changed in 001.submit_dock.sh :
condition="Unique name given to each experiment output - otherwise 'Default' "
seed="Random seed - otherwise '0' "
Submit the job after specifying partition and wall clock criteria. Typically ~2 minutes per system per core is sufficient.
sbatch 001.submit_dock.sh
This calls the script FLX.sh for each system which writes a dock input file and then immediately calls DOCK6.
A separate dock input file is written for each system. Below is the input file for DOCK6.10, but best practice would be to develop an input file in FLX.sh by first interactively creating an input file with the version of DOCK being used. This will prevent any changes in queries being overlooked:
conformer_search_type flex
write_fragment_libraries no
user_specified_anchor no
limit_max_anchors no
min_anchor_size 5
pruning_use_clustering yes
pruning_max_orients 1000
pruning_clustering_cutoff 100
pruning_conformer_score_cutoff 100.0
pruning_conformer_score_scaling_factor 1.0
use_clash_overlap no
write_growth_tree no
use_internal_energy yes
internal_energy_rep_exp 12
internal_energy_cutoff 100.0
ligand_atom_file /${testset}/${system}/${system}.lig.gast.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd yes
use_rmsd_reference_mol yes
rmsd_reference_filename /${testset}/${system}/${system}.lig.gast.mol2
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file /${testset}/${system}/${system}.rec.clust.close.sph
max_orientations 1000
critical_points no
chemical_matching no
use_ligand_spheres no
bump_filter no
score_molecules yes
contact_score_primary no
grid_score_primary yes
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix /${testset}/${system}/${system}.rec
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
minimize_flexible_growth_ramp yes
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_initial_score_coverge 5
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_anchor_max_iterations 500
simplex_grow_max_iterations 250
simplex_grow_tors_premin_iterations 0
simplex_random_seed $seed
simplex_restraint_min no
atom_model all
vdw_defn_file
/gpfs/projects/rizzo/ccorbo/DOCK_Builds/dock6.10_mpi/parameters/vdw_AMBER_parm99.defn
flex_defn_file
/gpfs/projects/rizzo/ccorbo/DOCK_Builds/dock6.10_mpi/parameters/flex.defn
flex_drive_file
/gpfs/projects/rizzo/ccorbo/DOCK_Builds/dock6.10_mpi/parameters/flex_drive.tbl
ligand_outfile_prefix ${condition}_$seed
write_orientations no
num_scored_conformers 1000
rank_ligands no
When docking is completed you will have a separate directory for each system. In each directory will be the input file, output file, and mol2 of docked results. If condition was "Default" and seed was "0", the file will be named:
${pdb_id}/Default_0_scored.mol2
IV.Pose Reproduction Analysis
Next run a script which calculates outcomes. This script is compatible with python 2.
module load py/2.7.15
python calculate_dock6.results.py ${condition}_${seed} ${system_file}
e.g.: python calculate_dock6.results.py Default_0 clean.systems.all
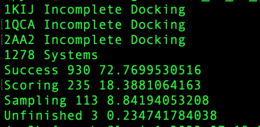
In the below image, the systems "1KIJ","1QCA","2AA2" did not successfully dock. There were 1,279 systems in the list provided.
-SEE README FILE IN GIT REPO FOR ADDTIONAL DETAILS THAT MAY NOT BE COVERED HERE
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (2024)
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>