2016 DOCK tutorial with Beta Trypsin
From Rizzo_Lab
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2014, using DOCK v6.6.
Contents
I. Introduction
Yaping She & Omar Sanchez Reyes
DOCK
Beta Trypsin
Organizing Directories
II. Preparing the Receptor and Ligand
Beilei Jiang
III. Generating Receptor Surface and Spheres
Lauren Prentis
Generating the Receptor Surface
Placing Spheres
IV. Generating Box and Grid
Monaf Awwa
Box Generation
Grid computing
V. Docking a Single Molecule for Pose Reproduction
Agatha Lyczek & Haoyue Guo
Minimization
Dock Minimization Input file: min.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file ../zzz.parameters/vdw.defn flex_defn_file ../zzz.parameters/flex.defn flex_drive_file ../zzz.parameters/flex_drive.tbl ligand_outfile_prefix 1BJU.min write_orientations no num_scored_conformers 1 rank_ligands no
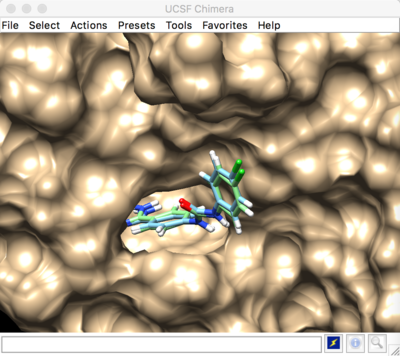
Result:
Rigid docking
To run the input file:
dock6 -i rigid_nr.in
Rigid docking input file: rigid_nr.in
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphere/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes atom_model all vdw_defn_file ../zzz.parameters/vdw.defn flex_defn_file ../zzz.parameters/flex.defn flex_drive_file ../zzz.parameters/flex_drive.tbl ligand_outfile_prefix 1BJU.rigid_nr write_orientations no
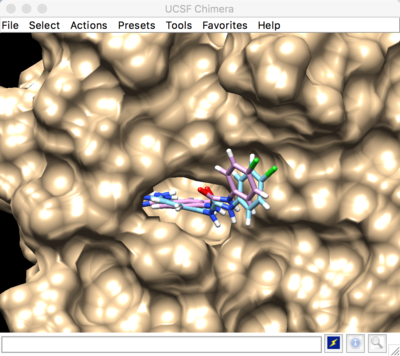
Best scored rigid docking result:
Flexible Docking
To run the input file:
dock6 -i flex.in
Flexible docking input file: flex.in
conformer_search_type flex user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 1000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 ligand_atom_file ../01.dockprep/1BJU.lig.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../02.surface-sphere/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../03.box-grid/1BJU.grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no ph4_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_descriptor_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0
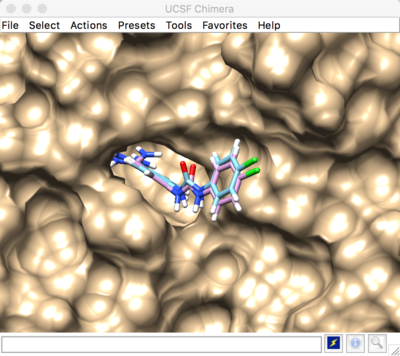
Result:
VI. Virtual Screening
Katie Maffucci
DOCK Preparation
Post Processing
VIII. Frequently Encountered Problems
EVERYONE