2016 DOCK tutorial with Beta Trypsin
For additional Rizzo Lab tutorials see DOCK Tutorials. Use this link Wiki Formatting as a reference for editing the wiki. This tutorial was developed collaboratively by the AMS 536 class of 2014, using DOCK v6.6.
Contents
I. Introduction
Yaping She & Omar Sanchez Reyes
DOCK
Beta Trypsin
Organizing Directories
<code>While performing docking, it is convenient to adopt a standard directory </code> <code>structure / naming scheme, so that files are easy to find / identify.</code> <code> For this tutorial, we will use something similar to the following:</code>
~username/AMS536-Spring2016/dock-tutorial/00.files/
:/01.dockprep/
:/02.surface-spheres/
:/03.box-grid/
:/04.dock/
:/05.large-virtual-screen/
:/06.virtual-screen/
:/07.footprint/
:/08.print_fps<pre>
<pre><ttIn addition, most of the important files that are derived from the original</tt>
crystal structure will be given a prefix that is the same as the PDB code, '1BJU'. The following
sections in this tutorial will adhere to this directory structure/naming scheme.
II. Preparing the Receptor and Ligand
Download the PDB File (1BJU)
Go to the PDB website to download 1BJU.pdb file(PDB code: 1BJU).
Using command or WinSCP to transfer 1BJU.pdb to 00.files. If you are in 00.files now, the command is:
cp ~/Downloads/1BJU.pdb .
Edit the PDB File (1BJU)
Using vi command to open 1BJU.pdb, then deleting bottom and top lines which do not have ATOM or ATPATM in its start.
Then changing all ATPATM to ATOM using command:
%s /ATPATM/ATOM /gc
In addition, changing all GP6 to LIG B using command:
%s /GP6 /LIG B /gc
Saving this as raw_1BJU.pdb in 00.files.
Prepare ligand and receptor files for dock
Create dockprep file
Open raw_1BJU.pdb file in chimera, in Tools, find structure editing, then click AddH to add hydrogen in this system.
Also in structure editing, click Add charge, then, changing AMBER ff14SB to AMBER ff99SB and changing net charge to +1.
Saving it as 1BJU.dockprep.mol2 in 01.dockprep.
Create receptor file
Open 1BJU.dockprep.mol2 in chimera, then click select to choose ligand, click action in Atoms/Bonds click delete.
Saving this molecule as 1BJU.rec.mol2 in 01.dockprep.
Create ligand file
Open 1BJU.dockprep.mol2 in chimera, using the same method delete the protein molecule.
Saving it as 1BJU.lig.mol2 in 01.dockprep.
Create no hydrogen receptor file
Open 1BJU.rec.mol2 in chimera, click select, then choosing chemistry H to select hydrogen in protein, Using action button to delete H.
Saving it as 1BJU.rec.noH.pdb in 01.dockprep.
III. Generating Receptor Surface and Spheres
Lauren Prentis
Generating the Receptor Surface
Placing Spheres
IV. Generating Box and Grid
Monaf Awwa
Box Generation
>Start Chimera and open the RECEPTOR files 1BJU.rec.noH.pdb
Next, select the ACTION subheader, then select SURFACE, then SHOW
Once you can see the van der Waals surface of the protein, select the
TOOLS subheader, then STRUCTURE EDITING, and lastly select WRITE DMS and save the file as 1BJU.dms
Now that we have a surface, we can more effectively calculate regions of the protein where a small molecule can bind. To further accelerate calculations, we will specify which atoms to calculate the interactions for via the program sphgen
Switch over to the 02.surfacespheres directory with the following command
cd 02.surfacespheres
Next, create an input file for sphgen to read.
vim INSPH
The file should have the following information
1BJU.rec.dms #the file corresponding to the surface is the input file
R #we want the spheres outside the protein surface
X #we want all subsets of spheres to be generated
0.0 #default size of proteins with surface contacts
4.0 #default maximum of the sphere radius generated (Angstroms)
1.4 #default minimum of the sphere radius generated (Angstroms) Also corresponds to probe radius
1BJU.rec.sph #output file name
Now we can enter the command to run sphgen
sphgen -i INSPH -o OUTSPH
Once completed, check the OUTSPH file to ensure no errors occured
If you'd like to visually inspect the spheres generated, start Chimera and load the 1BJU.rec.mol2 file, then load the 1BJU.rec.sph file.
Now we will use the showsphere program to convert our .sph file back to a .pdb format.
In the same directory, enter the command
showsphere
The following questions will appear. Answer as follows
Enter name of sphere cluster file:
1BJU.rec.sph
Enter cluster number to process (<0 = all):
-1
Generate surfaces as well as pdb files (<N>/Y)?
N
Enter name for output file prefix:
output_spheres
Process cluster 0 (contains ALL spheres) (<N>/Y)?
N
You should now have a file named output_spheres.pdb in your directory. You can visualize the spheres by loading chimera and the 1BJU.rec.noH.mol2, then loading the output_spheres.pdb file.
For the docking calculation, we are most interesting in a subset of the spheres. To select the spheres most important to ligand binding, we will use the command
sphere_selector 1BJU.rec.sph ../01.dockprep/1BJU.lig.mol2 8.0
The 8.0 Angstrom value indicates we want to select spheres within 8 angstroms of our ligand. This hypothetically means we are designing a competitive inhibitor.
Now lets create the proper output file for our spheres. Enter the command
showsphere
and answer the following questions
Enter name of sphere cluster file:
selected_spheres.sph
Enter cluster number to process (<0 = all):
-1
Generate surfaces as well as pdb files (<N>/Y)?
N
Enter name for output file prefix:
output_spheres_selected
Process cluster 0 (contains ALL spheres) (<N>/Y)?
N
This will ensure the docking calculation samples the most important regions of the protein surface
Grid computing
Since the electronic interactions between two objects in space can be decomposed into pairwise interactions, it is highly efficient to fully calculate a proteins electric contribution to binding energy before DOCKing a ligand.
switch to the 03.box-grid/ directory
cd 03.box-grid/
create an input file named showbox.in
vim showbox.in
enter the following information
Y # Do you want to calculate a box? (YES)
8.0 # How long should the box length be?
../02.surface-spheres/selected_spheres.sph # which sphere file is our input?
1 # How many clusters for the file?
1BJU.box.pdb # What do we want to name the output file?
save the file, then enter the following command to generate the box
showbox > showbox.in
The box can be visualized in Chimera by loading the output_spheres_selected.pdb file, then loading the 1BJU.box.pdb file
Next, we will calculate the grid energies
Create the input file for grid. Enter the command
vim grid.in
Run grid with the command
grid - grid.in
This allows you to go back and fix the input file in case there is a mistake when answer the questions prompted by grid.
grid.in
Parameter Value Description
compute_grids yes we want to compute grids
grid_spacing 0.4 resolution of grid energetic contributions (Angstroms). The smaller the value, the more accurate the calculation
output_molecule no we don't want to rewrite ligand output file
contact_score no we don't want to deal with contact score
energy_score yes we want to compute energetic interaction based on force field
energy_cutoff_distance 9999 what distance should we stop computing atomic interactions in angstroms
atom_model a corresponds to all atom model rather than united atom
attractive_exponent 6 see Lennard-Jones attraction term
repulsive_exponent 12 see Lennard-Jones repulsion term
distance_dielectric yes linear decrease in electric force
dielectric_factor 4 how much will electric force decrease based on atomic distance in angstroms
bump_filter yes
bump_overlap 0.75
receptor_file ../01.dockprep/1BJU.rec.mol2
box_file 4TKG.box.pdb
vdw_definition_file ../zzz.parameters/vdw_AMBER_parm99.defn characteristic van der Waal radii parameter file
score_grid_prefix grid ONLY THE PREFIX OF THE GRID FILE NAME
V. Docking a Single Molecule for Pose Reproduction
Agatha Lyczek & Haoyue Guo
Minimization
Dock Minimization Input file: min.in
conformer_search_type rigid
use_internal_energy yes
internal_energy_rep_exp 12
ligand_atom_file ../01.dockprep/1BJU.lig.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd yes
use_rmsd_reference_mol yes
rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2
use_database_filter no
orient_ligand no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../03.box-grid/1BJU.grid
multigrid_score_secondary no
dock3.5_score_secondary no
continuous_score_secondary no
footprint_similarity_score_secondary no
ph4_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
simplex_max_iterations 1000
simplex_tors_premin_iterations 0
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_random_seed 0
simplex_restraint_min yes
simplex_coefficient_restraint 10.0
atom_model all
vdw_defn_file ../zzz.parameters/vdw.defn
flex_defn_file ../zzz.parameters/flex.defn
flex_drive_file ../zzz.parameters/flex_drive.tbl
ligand_outfile_prefix 1BJU.min
write_orientations no
num_scored_conformers 1
rank_ligands no
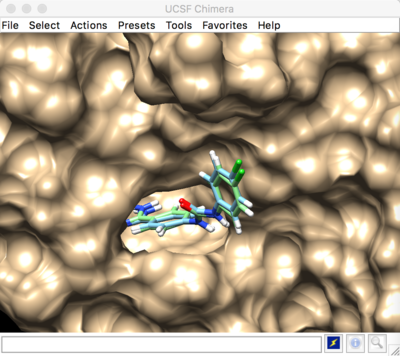
Result:
Rigid docking
To run the input file:
dock6 -i rigid_nr.in
Rigid docking input file: rigid_nr.in
conformer_search_type rigid
use_internal_energy yes
internal_energy_rep_exp 12
ligand_atom_file ../01.dockprep/1BJU.lig.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd yes
use_rmsd_reference_mol yes
rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file ../02.surface-sphere/selected_spheres.sph
max_orientations 1000
critical_points no
chemical_matching no
use_ligand_spheres no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../03.box-grid/1BJU.grid
multigrid_score_secondary no
dock3.5_score_secondary no
continuous_score_secondary no
footprint_similarity_score_secondary no
ph4_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
simplex_max_iterations 1000
simplex_tors_premin_iterations 0
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
simplex_rot_step 0.1
simplex_tors_step 10.0
simplex_random_seed 0
simplex_restraint_min yes
atom_model all
vdw_defn_file ../zzz.parameters/vdw.defn
flex_defn_file ../zzz.parameters/flex.defn
flex_drive_file ../zzz.parameters/flex_drive.tbl
ligand_outfile_prefix 1BJU.rigid_nr
write_orientations no
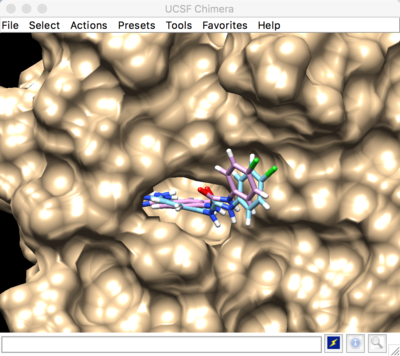
Best scored rigid docking result:
Flexible Docking
To run the input file:
dock6 -i flex.in
Flexible docking input file: flex.in
conformer_search_type flex
user_specified_anchor no
limit_max_anchors no
min_anchor_size 5
pruning_use_clustering yes
pruning_max_orients 1000
pruning_clustering_cutoff 100
pruning_conformer_score_cutoff 100.0
use_clash_overlap no
write_growth_tree no
use_internal_energy yes
internal_energy_rep_exp 12
ligand_atom_file ../01.dockprep/1BJU.lig.mol2
limit_max_ligands no
skip_molecule no
read_mol_solvation no
calculate_rmsd yes
use_rmsd_reference_mol yes
rmsd_reference_filename ../01.dockprep/1BJU.lig.mol2
use_database_filter no
orient_ligand yes
automated_matching yes
receptor_site_file ../02.surface-sphere/selected_spheres.sph
max_orientations 1000
critical_points no
chemical_matching no
use_ligand_spheres no
bump_filter no
score_molecules yes
contact_score_primary no
contact_score_secondary no
grid_score_primary yes
grid_score_secondary no
grid_score_rep_rad_scale 1
grid_score_vdw_scale 1
grid_score_es_scale 1
grid_score_grid_prefix ../03.box-grid/1BJU.grid
multigrid_score_secondary no
dock3.5_score_secondary no
continuous_score_secondary no
footprint_similarity_score_secondary no
ph4_score_secondary no
descriptor_score_secondary no
gbsa_zou_score_secondary no
gbsa_hawkins_score_secondary no
SASA_descriptor_score_secondary no
amber_score_secondary no
minimize_ligand yes
minimize_anchor yes
minimize_flexible_growth yes
use_advanced_simplex_parameters no
simplex_max_cycles 1
simplex_score_converge 0.1
simplex_cycle_converge 1.0
simplex_trans_step 1.0
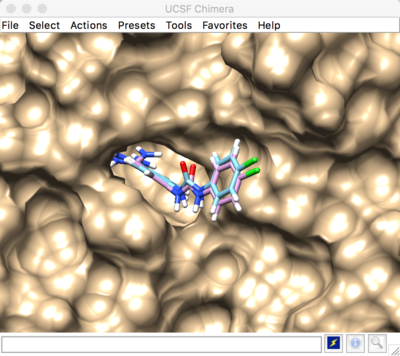
Best scored flexible docking result:
VI. Virtual Screening
Katie Maffucci
DOCK Preparation
Post Processing
VIII. Frequently Encountered Problems
EVERYONE