2019 Covalent docking tutorial 1 with PDB 2VKG
This tutorial teaches you how to dock a covalently bound drug molecule to a receptor (PDB 2VKG).
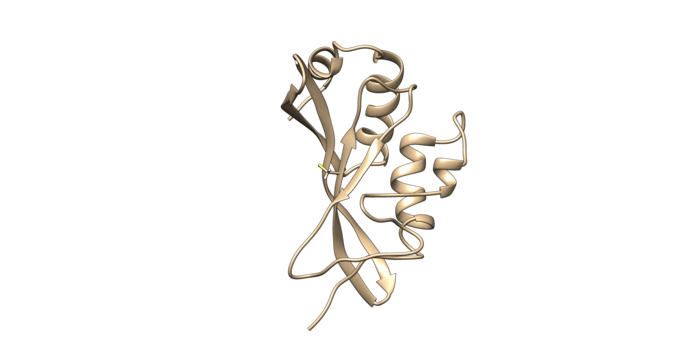
Use chimera to visualize your system in this case the pdb 2VKG. Chimera will not show a covalent linker between two separate entities, but if you check the distance you will notice the distance from the two sulfurs is about 2 angstroms.
I. Receptor Preparation
First step is to prepare the receptor. To do this open up the original pdb of the molecule. To prepare the receptor, you need to cut off the covalent linkers to generate the receptor. The newly formed receptor is now a neutral cysteine. Now charge the protein with a full neutral cysteine residue. To make the cysteine neutral, protons will be added into the system. Protonate with the latest AMBER force field, currently AMBER14SB.
To prepare it for making a grid, delete the side chain of the covalently bonded cysteine residue. When deleting the side chain of the residue make sure to delete the beta carbon but leave the alpha carbon. That is the receptor that will be used for docking and for the dock grid.
II. Ligand Preparation
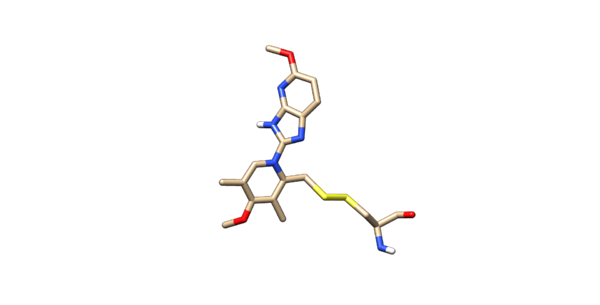
Open of the original structure. Delete all the protein leaving only the ligand covalently bonded too the cystine, leave the cystine attached to it. The ligand should be binded to the side chain residue all the way up to the beta carbon. Protonate the modified ligand and charge it with AM1BCC. Then delete all of the hydrogens off the beta-carbon. Modify the beta carbon so the name of the residue is D1. Chain or modify the sulfur atom so the name is D2. In VI change the atom type of the beta carbon and the sulfur to Du. The atoms type are being changed to dummy atoms. This is the end of the ligand preparation
Manually edit the mol2 so that the two sulfurs are connected. Short example shown below:
@<TRIPOS>ATOM
6 SG -24.3290 32.6070 -37.1160 S.4 1 CYS 0.0000
32 S20 -24.6380 34.0060 -37.3390 S.3 1 LIG 0.0000
@<TRIPOS>BOND
1 1 7 1
2 6 32 1
The @<TRIPOS>ATOM denotes the coordinates of the atoms which should stay unchanged. The @<TRIPOS>BOND denotes the connectivity records in the mol2. You will need to add another connection between the two sulfurs. The first columns is the bond number. The second column is the atom number of the atom seen first (the lowest atom number goes first). The third column is the atom number of the second sulfur (the higher atom number). The fourth and final column is the bond type, 1 being a single bond. Highlighted in bold above shows the modified bond connectivity. The ligand should look connected to the cysteine residue as seen below.
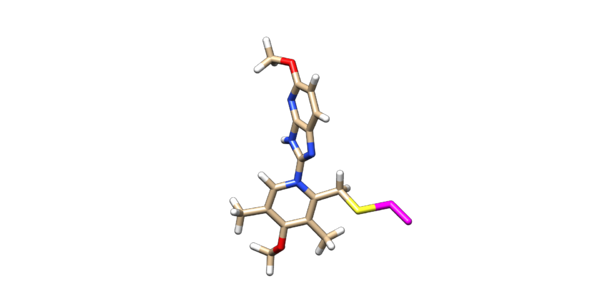
Delete the backbone of the cysteine leaving the beta carbon. Protonate and charge the ligand using AM1BCC. After the ligand is protonated and charged, delete the hydrogen's off of the beta carbon. Then manually change the atom name of the beta carbon to D2 and change the atom type from C.3 to Du. Change the atom name of the sulfur connected to the beta carbon to D1 and changed the atom type to Du as seen below:
@<TRIPOS>ATOM
1 D2 -24.0727 34.9285 -36.1952 Du 1 LIG -0.0660
2 D1 -25.1580 36.1840 -35.7130 Du 1 LIG -0.1111
The ligand should now look like this in chimera:
III. Preparing the sphere files for orienting
The only spheres that will be orientated will be the alpha carbon, beta carbon, and the sulfur on the cysteine sidechain. Isolate the sulfur, alpha and beta carbons and delete everything else. Once that is isolated save it as a pdb or as a mol2. Convert this pdb or mol2 into a sphere file manually. Save the sphere file for orienting when docking.
Now new spheres will be generated for the box. To accomplish this spheres will be generated ligand file. A sphere file will be made using the coordinates from the ligand file of the heavy atom. Now that the spheres are generated twice once for orientating and the other for the dock/grid. The box will then be generated, once the box is made the grid can be populated in the box. Now that the grid, the ligand, and spheres meant of orientating. You can begin covalent docking.