2022 DOCK tutorial 3 with PDBID 1X70
Contents
Introduction
DOCK
System Preparation
Fetching 1X70
Open Chimera and do the following to grab the protein:
File > Fetch By ID > 1X70
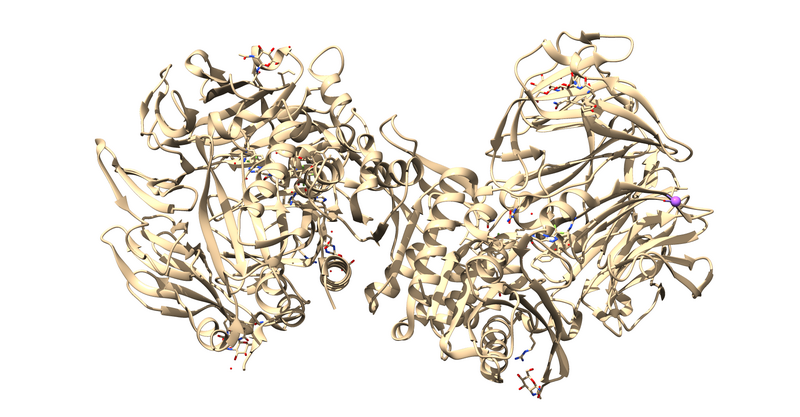
The first thing to notice is that this is a dimer and the ligand, 715, is not bound at the dimer interface. Thus, one of the monomers is entirely redundant and should be deleted.
Select > Chain > A
Actions > Atoms > Delete
Next it is important to remove cofactors, ions, and water molecules not involved in the binding interactions. This can be checked by reading through the paper associated with the PDB.
Select > Residue > NAG > Actions > Atoms > Delete
Select > Residue > NDG > Actions > Atoms > Delete
Select > Residue > HOH > Actions > Atoms > Delete
This will leave us with just the ligand and receptor.
Receptor Prep
Now that we the receptor by itself, we have to clean up the rest of the receptor by adding any missing side chains, dealing with multiple occupancies and mutated residues, and protonating and calculating partial charges.
For 1X70, there are several residues with multiple occupancies.
One way to check for these residues is to grep the number of alpha carbons from the pdb in the command line.
In the terminal, type: grep -e CA 1X70_noH.pdb
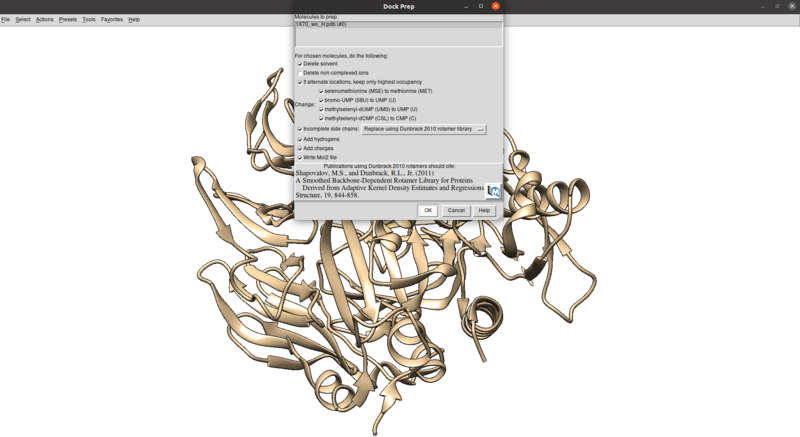
To clean this up and protonate/charge the receptor:
Tools > Structure Editing > Dock Prep
Make sure that the protonation makes sense for residues in the active site or coordinated with metals (none here), especially histidines, by checking the paper and chimera for any nitrogens coordinating with metals are not protonated or residues in the active site with differing protonation states.
Ligand Prep
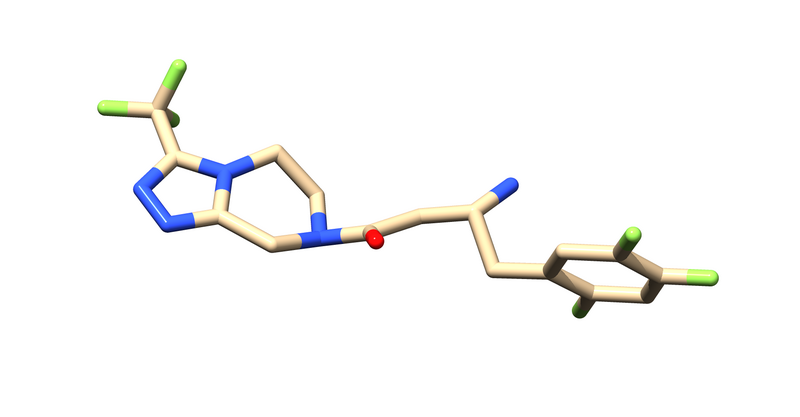
First you have to save the ligand as a separate file. You can do this in Chimera by deleting all of the protein and saving that as a separate file: "715_noH.pdb".
Select > Residues > 715 > Invert Selection Actions > Atoms > delete File > save PDB
Now you should just have the ligand.
Next Hydrogens and partial atomic charges need to be added and saved as "715_H.mol2". It will ask for the ligands overall charge, which you should verify using chemical knowledge.
tools > structure editing > addH tools > structure editing > addCharge > Select the correct charge for your ligand, Use AM1-BCC. File > save Mol2
Surface Generation & Spheres
This section details the generation of sphere files which will be used to describe where you are trying to DOCK to on your protein.
Surface Generation
Load 1X70 w/o Hydrogens in Chimera actions > show > surface
Then you will write a DMS (Molecular Surface) File With the surface generated in Chimera: Tools > Structure Editing > Write DMS
Now you should have a DMS file for the next step.
Sphere Generation To generate spheres make the following input file: "INSPH" -
1X70_dms.dms #Molecular Surface File R #Whether to generate spheres outside of surface (R) or inside (L) X #Surface points from the DMS file to use in sphere generation 0 #Minimum radius between spheres 4.0 #Maximum radius of sphere 1.4 #Minimum radius of sphere 1X70_wo_H.sph #Output sphere file
For more information on sphere generation see: https://dock.compbio.ucsf.edu/DOCK_6/tutorials/sphere_generation/generating_spheres.htm
The .sph file should give you something similar to the following image if you load it up over your protein in Chimera:
Sphere Selection
Making The Infamous Grid
Making the box
In order to make the grid we first have to determine how big our grid will be. To do this:
cd 03.grid
showbox
#automatically construct box to enclose spheres [Y/N]
Y
#extra margin to also be enclosed (angstroms)?
#(this will be added in all 6 directions)
8.0
#sphere file-
./../02.spheres/selected_spheres.sph
#cluster number-
1
#output filename?
1X70.box.pdb
You can also copy these inputs into "showbox.in" (none of the commented lines) and then type "showbox < showbox.in"
Making the grid
Energy Minimization for the ligand
Docking & Virtual Screening
Rigid Docking
Fixed Anchor Docking
Flexible Docking
Placeholder
[[File:|thumb|center|800px|image placeholder]]