File list
This special page shows all uploaded files.
First page |
Previous page |
Next page |
Last page |
| Date | Name | Thumbnail | Size | User | Description | Versions |
|---|---|---|---|---|---|---|
| 11:07, 27 February 2012 | DOCK 3rd run result.png (file) |  |
301 KB | Stonybrook | 1 | |
| 11:04, 27 February 2012 | 2012 DOCK Tutorial 1DF8 dms.png (file) |  |
50 KB | Stonybrook | Reverted to version as of 15:01, 27 February 2012 | 4 |
| 10:55, 27 February 2012 | VirtualScreeningTest.png (file) |  |
50 KB | Stonybrook | 1 | |
| 10:50, 27 February 2012 | DOCK 2nd run result.png (file) |  |
301 KB | Stonybrook | By only setting orient_ligand option to "yes", the docking result looks pretty bad. | 1 |
| 10:48, 27 February 2012 | 2012 DOCK Tutorial 1DF8 surface.png (file) |  |
172 KB | Stonybrook | 1 | |
| 18:02, 26 February 2012 | 1DF8 box.png (file) |  |
141 KB | Stonybrook | Chimera rendering of 1DF8 receptor with a ligand and the box generated around the ligand using showbox program. | 1 |
| 15:20, 26 February 2012 | Flow chart.JPG (file) |  |
63 KB | Stonybrook | Flow Chart of Questions for Showbox (Red path is followed in this tutorial) source:http://dock.compbio.ucsf.edu/DOCK_6/tutorials/grid_generation/generating_grid.html | 1 |
| 05:55, 7 February 2012 | GP41 research image1.png (file) |  |
76 KB | WikiSysop | 1 | |
| 13:02, 3 February 2012 | ErbB Kinases research image1.png (file) |  |
44 KB | Tbalius | 1 | |
| 12:28, 3 February 2012 | FPS Dock Scoring research image1.png (file) |  |
58 KB | Tbalius | 1 | |
| 16:57, 6 December 2011 | Ubq ss.jpg (file) |  |
70 KB | Stonybrook | 3 | |
| 16:56, 6 December 2011 | Ubq scount.jpg (file) |  |
57 KB | Stonybrook | 4 | |
| 16:56, 6 December 2011 | Ubq rmsf plot.jpg (file) |  |
27 KB | Stonybrook | 2 | |
| 16:56, 6 December 2011 | Ubq nvt energy.jpg (file) |  |
20 KB | Stonybrook | 2 | |
| 16:56, 6 December 2011 | Ubq npt energy.jpg (file) |  |
38 KB | Stonybrook | 2 | |
| 16:52, 6 December 2011 | Ubq energy.jpg (file) |  |
20 KB | Stonybrook | 2 | |
| 16:49, 6 December 2011 | Ubq rmsd.jpg (file) |  |
36 KB | Stonybrook | 3 | |
| 11:05, 27 July 2011 | Ubq rmsf structure.jpg (file) |  |
24 KB | Stonybrook | 2 | |
| 15:13, 21 July 2011 | Ubq ions.jpg (file) |  |
65 KB | Stonybrook | 1 | |
| 13:22, 21 July 2011 | Ubq sol.jpg (file) |  |
79 KB | Stonybrook | 2 | |
| 12:54, 21 July 2011 | Pbc overlap.jpg (file) |  |
55 KB | Stonybrook | 2 | |
| 11:50, 15 June 2011 | Brian email.png (file) | 7 KB | Stonybrook | 1 | ||
| 15:36, 1 June 2011 | Joe allen email.png (file) | 7 KB | Stonybrook | 1 | ||
| 21:28, 23 May 2011 | DNA.png (file) |  |
5 KB | Stonybrook | 1 | |
| 21:26, 23 May 2011 | DNA.jpg (file) |  |
5 KB | Stonybrook | 2 | |
| 10:18, 19 May 2011 | Gleevec.correct.png (file) |  |
66 KB | Tbalius | 1 | |
| 17:47, 9 March 2011 | Selected spheres.dock tutorial.2011.png (file) |  |
364 KB | Sudipto | 1 | |
| 17:25, 9 March 2011 | All spheres.dock tutorial.2011.png (file) |  |
443 KB | Sudipto | 1 | |
| 17:42, 2 March 2011 | RMSD Plot (file) |  |
29 KB | Stonybrook | 2 | |
| 17:39, 2 March 2011 | RMSD plot (file) | 121 KB | Stonybrook | 1 | ||
| 14:22, 9 December 2010 | 2010.ams535.class.picture.jpg (file) |  |
186 KB | Rizzo | 2 | |
| 18:16, 13 September 2010 | CARBONIC ANHYDRASExtalmin.1000iter-tether10ENERGYbw.png (file) | Error creating thumbnail: File with dimensions greater than 12.5 MP |
324 KB | Sudipto | 1 | |
| 15:13, 19 August 2010 | Lingling.email.png (file) | 678 bytes | Lingling | 1 | ||
| 18:05, 29 June 2010 | Jibril.email.png (file) | 1 KB | Stonybrook | 3 | ||
| 13:58, 10 April 2010 | E asis (file) | 6 KB | Kehauser | Use Word Pad to view in windows. This is the Perl script from the Amber installation. It could also be found in the Amber Tutorials. "Process_mdout.pl" is the filename, I believe.. | 1 | |
| 15:40, 10 March 2010 | 111.png (file) |  |
22 KB | Eric | 1 | |
| 23:45, 8 March 2010 | Graph1.jpg (file) | 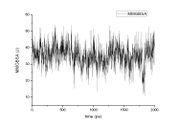 |
238 KB | Haoquan | 1 | |
| 23:19, 8 March 2010 | Data.MMGBSA vs time (file) | 26 KB | Haoquan | 1 | ||
| 17:42, 5 March 2010 | 2009.ams535.class.picture.jpg (file) |  |
185 KB | Rizzo | 3 | |
| 17:35, 5 March 2010 | 2008.ams535.class.picture.jpg (file) |  |
65 KB | Rizzo | 1 | |
| 19:37, 1 March 2010 | Wikilogo.png (file) |  |
24 KB | Tbalius | 1 | |
| 16:12, 1 March 2010 | MMGBSA ΔΔG.txt (file) | 25 KB | Haoquan | 1 | ||
| 17:59, 22 February 2010 | 2nd best rigiddock.png (file) |  |
106 KB | Anchang | 1 | |
| 17:59, 22 February 2010 | Best rigiddock.png (file) |  |
104 KB | Anchang | 1 | |
| 10:40, 20 February 2010 | Surface.png (file) | 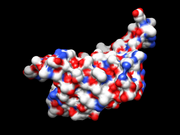 |
115 KB | Anchang | Streptavidin enzyme surface | 1 |
| 01:51, 12 February 2010 | T4 data result.jpg (file) | 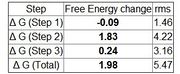 |
17 KB | Stonybrook | 1 | |
| 01:47, 12 February 2010 | T4 TIMD results.jpg (file) |  |
125 KB | Stonybrook | 1 | |
| 01:45, 12 February 2010 | 图片.JPG (file) |  |
125 KB | Stonybrook | 1 | |
| 14:43, 9 February 2010 | 1DF8.pdb.gz (file) | 49 KB | Kehauser | Biotin Streptavidin pdb.gz | 1 | |
| 14:32, 9 February 2010 | Amber10i-1- - Manual.pdf (file) | 1,016 KB | Kehauser | Amber 10 Manual | 1 |
First page |
Previous page |
Next page |
Last page |