2020 DOCK tutorial 1 with PDBID 3VJK
Contents
Introduction
DOCK
DOCK 6 is a molecular modeling software that is used for investigating ligand binding geometry and ligand interactions[1]. Consequently, its relevance in the field of drug discovery is clear. This program was initially developed by Dr. Irwin Kuntz and colleagues at the University of California San Francisco. A major feature of DOCK 6 is the search algorithm that is used: “anchor-and-grow”; this sets the software apart from its counterparts [2]. This method first identifies the rigid structure of a particular ligand--anchors-- then the program docks the ligand using its geometry. Following the docking, a partial conformational search is performed. In simple terms, the positions of the anchor are allowed independently however once a favored conformation is found it is retained. Once this step is completed, energy minimization is carried out Full details here.
3VJK
3VJK is the PDB code for the crystal structure of human dipeptidyl peptidase IV, also known as DPP-4, with MP-513, which is called Teneligliptin [3]. DPP-4 is a symmetrical dimer and has 729 residues per chain. The crystal has a resolution of 2.49 Å, a R-value of 0.279, and a R-free value of 0.225. To add on, the molecule in the crystal--Teneligliptin--has been approved for the treatment of type II Diabetes in Japan and has shown promising results in vivo [4].
Software packages
To follow this tutorial you will need to have the following programs installed:
- DOCK
- Chimera
This tutorial used Dock 6.9 & Chimera 1.13.1
At several points this tutorial will reference these programs as commands in a shell environment. The students who did this ran their programs on a UNIX (CoreOS or Ubuntu) server, although this process should generalize to your specific setup. For help, please reference available documentation.
Directory Organization
The following tutorial will use the organization of directories prepared below. This specific organization is not required but is recommended. The "mkdir" command will be employed which creates a new folder in which files can be saved. To navigate into a directory use the command "cd" followed by the directory name. To change to the directory the next level up, use the command "cd .." .
Within the Bash Shell environment:
mkdir 3VJK cd 3VJK mkdir 001.structure 002.surface_spheres 003.gridbox 004.dock 005.virtual_screen 006.virtual_screen_mpi 007.cartesianmin 008.rescore
All eight directories should be created now and this can be visually confirmed with the command "ls".
Receptor Preparation
Preparing the Structure for Docking
Downloading and Opening PDB File
Download the PDB Format file from the associated rcsb page here. This web page includes associated articles, files, and other meta data.
Download files -> PDB Format
This file provides information on the 3D orientation of the atoms within the protein and ligand as well as any co-factors (any other molecules present during the crystallization experiment, typically water and metal ions). The file can be opened up and manipulated in the program Chimera.
Open Chimera:
File -> Open -> (Location where you downloaded PDB file)
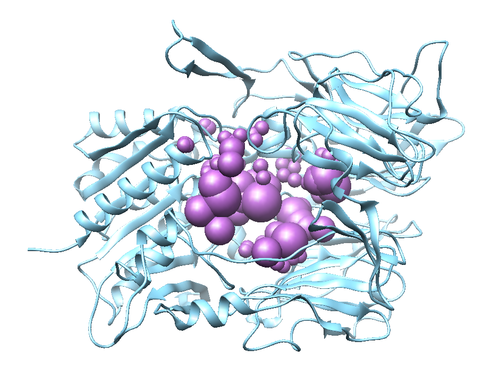
The protein should appear the same as the image above. The image can be rotated to view from different angles. This is called a Ribbon diagram and shows the backbone of the protein, however some amino acid side chains are shown by default. Also shown explicitly are NAG amino acid modifications, the Oxygen of several water molecules and M51 (the ligand that is complexed with the protein). There are no Hydrogen atoms represented anywhere. This is because PDB files do not contain information on Hydrogen atoms.
Preparation of the Protein Receptor for Docking
Docking requires that the protein receptor and ligand be separated into different files. First, the receptor file will be prepared. This particular protein is a homo-dimer (two identical units of the same peptide). For simplicity and to avoid possible complications in later steps, only one of the peptide chains will be retained. This step should be applied judiciously in protein systems where the ligand is at the interface of two dimers.
Select -> Chain -> B Actions -> Atoms/ Bonds -> Delete
Only one monomer of protein should remain now.
Next the NAG amino acid modifications, waters and ligand will be removed. They are not crucial for the Docking experiment, and may be problematic and cause failure if retained.
Select -> Residue -> All nonstandard Actions -> Atoms/ Bonds -> Delete
The receptor is now "clean" and should be saved prior to the next step.
File -> Save Mol2 -> "3VJK_rec_woH.mol2"
It is important to give files a logical naming scheme. The woH portion is to specify Hydrogens have not yet been added. Move this file to the directory "001.structure"
Adding Hydrogens and Charge
In order to calculate interactions between the protein and ligand, Hydrogens must be added to the receptor. Chimera will apply standard protonation states to the amino acids. It is important to check these protonation states afterwards, as they may not match the crystallization experiment. For example, the paper associated with the PDB being worked with may specify a certain residue is protonated. It would then be crucial to check this after the following step, and if it is incorrect, to adjust it manually.
Structure Editing -> Add H -> Ok
Next partial charges will be added to each atom in the receptor.
Structure Editing -> Add Charge -> (AM1BCC charges should be selected) -> Ok
Now save this as a mol2 file "3VJK_rec_dockprep.mol2" and move it to the directory "001.structure"
Ligand Preparation
Preparing Ligand We will now need to prepare the Ligand, M-513. In a similar manner to receptor preparation, open the PDB file on Chimera. Likewise, you will also need to delete Chain B as previously stated. Now, you will be able to isolate the ligand by doing the following:
select->residue->M-51 select->invert Actions->Atoms/Bonds->Delete
You should be left with the following:
Next, we will save this as a mol2 file:
File->save as mol2 ->3VJK_ligand_noH.mol2
Add Hydrogens and Charge The crystal structure does not have any hydrogens because of technical limitations; hydrogen electron densities are too small to be detected. Consequently, we must add hydrogens to the ligand.
Tools->Structure editing-> add H
In a similar fashion, DOCK will need charges to perform calculations.
Tools->Structure editing-> Add Charge
It is important to make a note about the net charge of the ligand. you should not assume that chimera has the correct charge''. You should look at the ligand and attempt to validate the charge, which should be +2. You can now save this as a mol2 file and name it: 3VJK_ligand_with_H.mol2
Surface Generation & Sphere Selection
Surface Generation In Chimera a file which represents the surface of the protein will be created. The surface will be used to create a negative image of the protein (spheres which occupy the cavities and external face of the protein). These spheres are used to guide the ligand during docking.
In Chimera open "3VJK_rec_woH.mol2" :
Actions -> Surface -> Show Tools -> Structure Editing -> Write DMS -> "3VJK_rec_surface.dms"
Move this to the directory "002.surface_spheres"
Sphere Selection
By this step, you should have the mol2 extractions of ligand and protein, in both hydrogenated and unhydrogenated forms (4 files). The next activity is to create an efficient representation of empty space inside the protein. This is done with the sphgen script, which tries to generate the largest possible sphere for any given empty space. In general, it is desirable for the spheres will eclipse with each other, but not with the protein itself.
The sphgen software takes in a series of inputs from prompts to the user, but we can automate this by piping these arguments through a file. We shall can this file INSPH. Generate your INSPH file with the following syntax:
[your_receptor].dms <R flag> - enables sphere generation outside the protein surface (no eclipsing) <X flag - uses all coordinates <double> - distance that steric interactions are checked (units?) <double> - Maximum sphere radius of generated sphere (units?) <double> - Size of sphere that rolls over dms file surface for cavities (units?) [your_receptor].sph
This is an example of how we wrote our file:
3VJK_rec_surface.dms R X 0.0 4.0 1.4 3VJK_receptor_woH.sph
Does it matter if the dms is generated with the hydrogens?
This should produce an sph file that you can then run through sphgen
sphgen -i INSPH -o OUTSPH
Sphere Selection
Using dock's sphere_selector script, we are able to produce a subset of spheres that are close (within 10 angstroms) to the ligand
sphere_selector 3vjk_receptor.sph 3vjk_ligand_H.mol2 10.0
Generation of the Box and Grid
Energy calculations can be computationally expensive. Consequently, steps must be performed in order to reduce the number of calculations that are performed. In more detail, DOCK will be calculating the energy using a grid. We will be generating the grid; anything that is beyond the grid generated will not be in the calculation. This means that will ignore long distant interactions with ligand.
To start, we will be making a directory for the grid and the box
mkdir 003.gridbox
Generating the box
Next, we will be creating an input file that contains information for the Showbox programs. This file will contain parameters for the box.
vi showbox.in
we will put the following in to the file:
Y #generate box# 8.0 #how many angstroms the box edges should be from the spheres# ./../002.surface_spheres/select_spheres.sph #the location of the selected spheres# 1 3VJK.box.pdb #name of the output file#
In order to run this you do this:
showbox < showbox.in
After you run this command a file called 3VJK.box.pdb will be generated. This files contains the grid. And can be visualized.
Generating the grid
In a similar manner, we will have to generate the grid. In order to do this we will need to make the input file for the grid program that contains:
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4. bump_filter yes bump_overlap 0.75 receptor_file d box_file d.box.pbd vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
we will called this file grid.in. In order to generate the grid do the following:
grid -i grid.in -o gridinfo.out
the "-o" flag is used to specify the name of the output file. Once the program is completed there should be three files generated: gridinfo.out, grid.nrg, and grid.bmp. It is a good idea to make sure that gridinfo.out matches with the known information of the system. In other words, this is a good spot to double check your work.