2022 DOCK tutorial 3 with PDBID 1X70
Contents
Introduction
DOCK
DOCK is a suite of software intended for molecular docking experiments. It was originally developed by Robert Sheridan, Renee DesJarlais, and Irwin Kuntz. It has been further developed by many additional authors and has many different versions and features. More information about DOCK or the features of DOCK6.9, the version used in this tutorial are available in the DOCK6 manual[1]
1X70
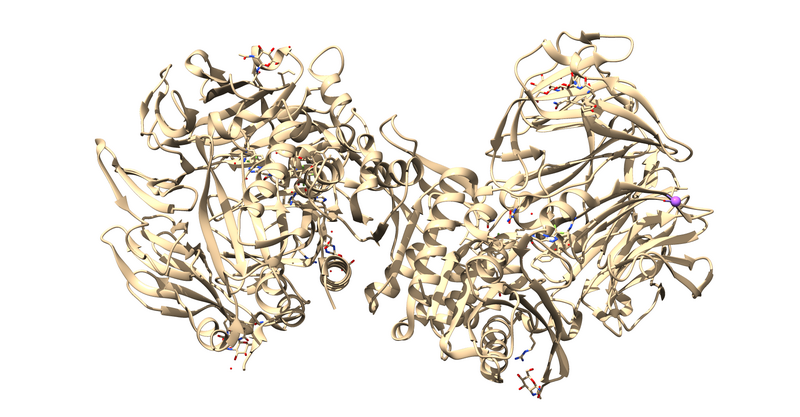
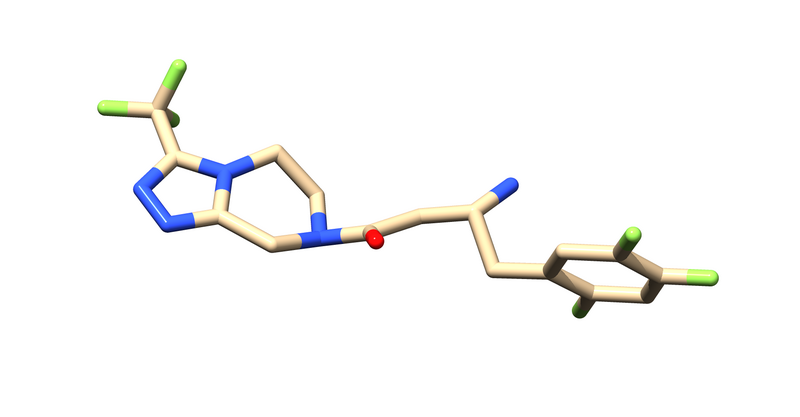
1X70 is a protease responsible for degrading a hormone peptide that has potential in mitigation the impact of type 2 diabetes[2]. It is a 174.94 kDa homodimer with 728 residues per monomer with ligand bound in both active sites[3]. It has a resolution of 2.1 Å with an R value of 0.193 and an R free value of 0.228[4].
System Preparation
Directory Setup Start by running the following command on two computers:
a computer which you can access the molecular visualization software Chimera a computer capable of running DOCK calculations:
mkdir 001.structure 002.spheres 003.gridbox 004.energy_min 005.footprint 006.vs 007.cart_min 008.rescore
IMPORTANT: Throughout this tutorial the paths of files and directories may vary. It is important to change them to match what you have on your local machine when preparing input files or your calculations will fail! Also consider this when shuttling files between computers for visualization using Chimera!
Fetching 1X70
Open Chimera and do the following to grab the protein:
File > Fetch By ID > 1X70
The first thing to notice is that this is a dimer and the ligand, 715, is not bound at the dimer interface. Thus, one of the monomers is entirely redundant and should be deleted.
Select > Chain > A
Actions > Atoms > Delete
Next it is important to remove cofactors, ions, and water molecules not involved in the binding interactions. This can be checked by reading through the paper associated with the PDB.
Select > Residue > NAG > Actions > Atoms > Delete
Select > Residue > NDG > Actions > Atoms > Delete
Select > Residue > HOH > Actions > Atoms > Delete
This will leave us with just the ligand and receptor.
Receptor Prep
Now that we the receptor by itself, we have to clean up the rest of the receptor by adding any missing side chains, dealing with multiple occupancies and mutated residues, and protonating and calculating partial charges.
For 1X70 one of the problems you will run into is that there are several residues with multiple occupancies.
One way to check for these residues is to grep the number of alpha carbons from the pdb in the command line.
In the terminal, type: grep -e CA 1X70_noH.pdb
To clean this up, avoid other potential issues including mutated residues and missing atoms/chains, and protonate/charge the receptor:
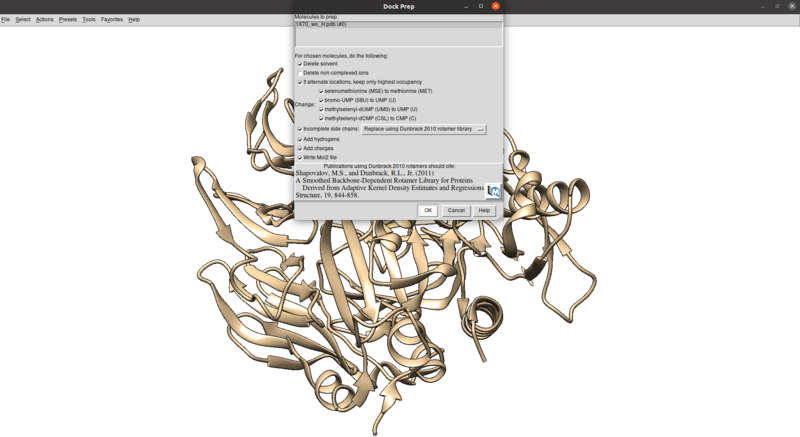
Tools > Structure Editing > Dock Prep
Make sure that the protonation makes sense for residues in the active site or coordinated with metals (none here), especially histidines, by checking the paper and chimera for any nitrogens coordinating with metals are not protonated or residues in the active site with differing protonation states. Also make sure that all residues have integral charge (i.e. the charge is an integer). Chimera should give a warning after the dockprep if any residue charges are non-integral.
Ligand Prep
First you have to save the ligand as a separate file. You can do this in Chimera by deleting all of the protein and saving that as a separate file: "715_noH.pdb".
Select > Residues > 715 > Invert Selection
Actions > Atoms > delete
File > save PDB
Now you should just have the ligand.
Next Hydrogens and partial atomic charges need to be added and saved as "715_H.mol2". It will ask for the ligands overall charge, which you should verify using chemical knowledge.
tools > structure editing > addH
tools > structure editing > addCharge > Select the correct charge for your ligand, Use AM1-BCC.
File > save Mol2
Surface Generation & Spheres
This section details the generation of sphere files which will be used to describe where you are trying to DOCK to on your protein.
Surface Generation
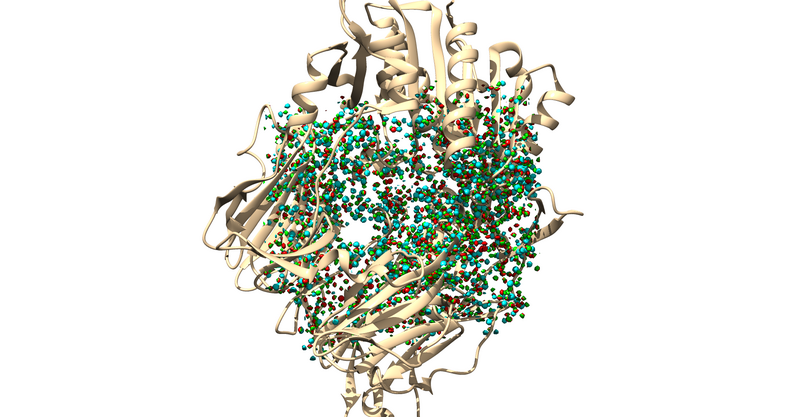
Load 1X70 w/o Hydrogens in Chimera
actions > show > surface
Then you will write a DMS (Molecular Surface) File With the surface generated in Chimera:
Tools > Structure Editing > Write DMS
Save this as 1X70_dms.dms, and now you should have a DMS file for the next step.
Sphere Generation To generate spheres make the following input file: "INSPH" -
1X70_dms.dms #Molecular Surface File R #Whether to generate spheres outside of surface (R) or inside (L) X #Surface points from the DMS file to use in sphere generation 0 #Minimum radius between spheres 4.0 #Maximum radius of sphere 1.4 #Minimum radius of sphere 1X70_wo_H.sph #Output sphere file
For more information on sphere generation see: https://dock.compbio.ucsf.edu/DOCK_6/tutorials/sphere_generation/generating_spheres.htm
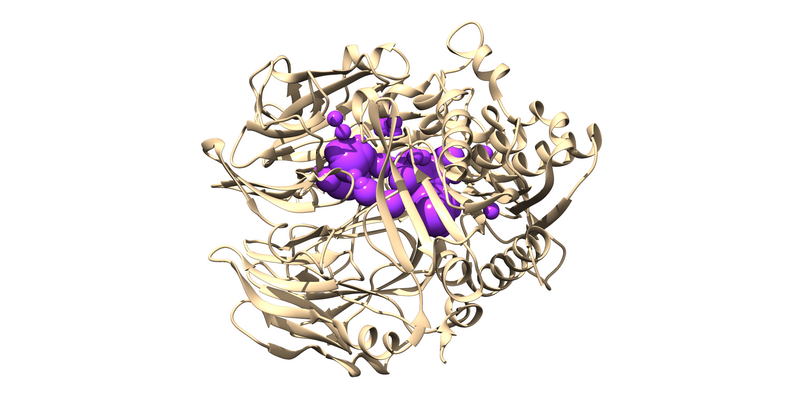
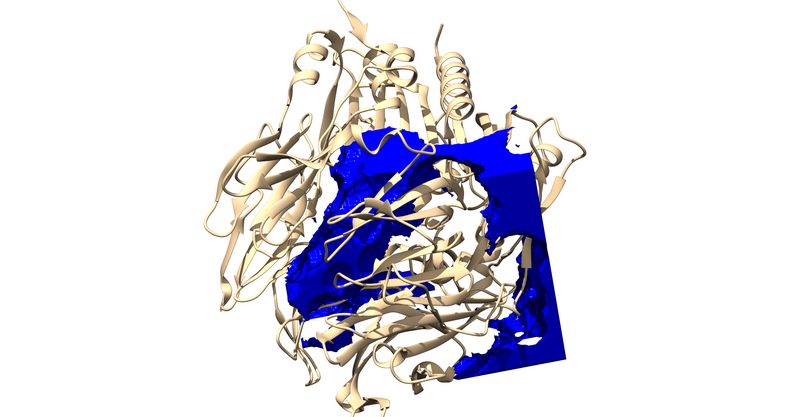
The .sph file should give you something similar to the following image if you load it up over your protein in Chimera:
Sphere Selection
Now we need to select the subset of all those spheres that will be used in the docking experiment by excluding those too far away from the ligand. We can do this through the following command:
#sphere_selector <receptor> <ligand> <radius in angstroms> sphere_selector 1X70_wo_H.sph 715_H.mol2 10.0
Making The Infamous Grid
Making the box
In order to make the grid we first have to determine how big our grid will be. To do this:
cd 03.grid
showbox
#automatically construct box to enclose spheres [Y/N]
Y
#extra margin to also be enclosed (angstroms)?
#(this will be added in all 6 directions)
8.0
#sphere file-
./../02.spheres/selected_spheres.sph
#cluster number-
1
#output filename?
1X70.box.pdb
You can also copy these inputs into "showbox.in" (none of the commented lines) and then type "showbox < showbox.in"
Making the grid
Making the following input file "grid.in":
compute_grids yes grid_spacing 0.4 output_molecule no contact_score no energy_score yes energy_cutoff_distance 9999 atom_model a attractive_exponent 6 repulsive_exponent 9 distance_dielectric yes dielectric_factor 4. bump_filter yes bump_overlap 0.75 receptor_file ../01.structures/1X70_rec.mol2 (this is the protonated and charged ligand) box_file ./1X70.box.pdb vdw_definition_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn score_grid_prefix grid
Now generate the grid. It should take several minutes.
grid -i grid.in -o gridinfo.out
Once it is done, vi into gridinfo.out and make sure the charges are all integer and that the calculation finished. If they are not, that likely means there is something wrong with your 1X70_H.mol2, and you may want to go back and double check your receptor prep step and remake this file.
You should get two files:
grid.nrg (energy grid) grid.bmp (bump grid)
The energy grid is used in the dock calculations to give you energies of interaction between the ligand and protein.
The bump grid is used to make sure the ligand doesn't overlap with any receptor atoms and also contains information of the other grids.
Try loading them into chimera over your protein.
Energy Minimization for the ligand
Now cd into directory 04.energy_min and make the following input file "min.in":
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ./../01.structures/715.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ./../01.structures/715.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ./../03.grid/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min yes simplex_coefficient_restraint 10.0 atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 1X70.lig.min write_orientations no num_scored_conformers 1 rank_ligands no
Then run this through dock
dock6 -i min.in -o min.out
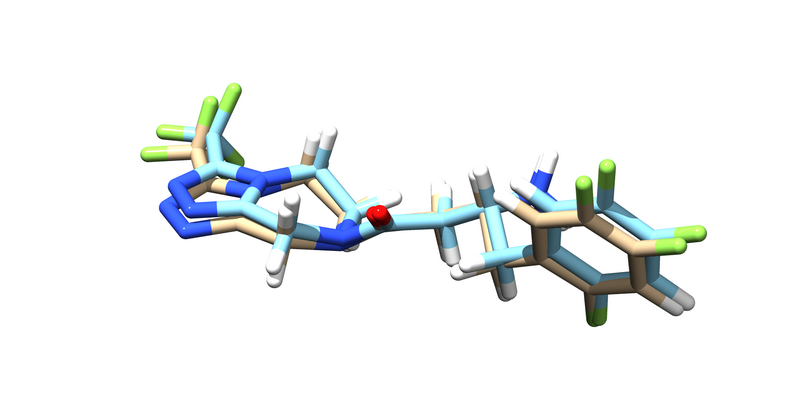
You should get an energy minimized version of the ligand. Try pulling it up over the original ligand in chimera.
Footprint Generation
Now if we wanted to see any changes between the interactions of the unminimized ligand and the minized pose, we can create the following file "footprint.in":
conformer_search_type rigid use_internal_energy no ligand_atom_file ./../04.energy_min/1X70.lig.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd no use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary no grid_score_secondary no multigrid_score_primary no multigrid_score_secondary no dock3.5_score_primary no dock3.5_score_secondary no continuous_score_primary no continuous_score_secondary no footprint_similarity_score_primary yes footprint_similarity_score_secondary no fps_score_use_footprint_reference_mol2 yes fps_score_footprint_reference_mol2_filename ./../01.structures/715.mol2 fps_score_foot_compare_type Euclidean fps_score_normalize_foot no fps_score_foot_comp_all_residue yes fps_score_receptor_filename ./../01.structures/1X70_rec.mol2 fps_score_vdw_att_exp 6 fps_score_vdw_rep_exp 9 fps_score_vdw_rep_rad_scale 1 fps_score_use_distance_dependent_dielectric yes fps_score_dielectric 4.0 fps_score_vdw_fp_scale 1 fps_score_es_fp_scale 1 fps_score_hb_fp_scale 0 pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix footprint.out write_footprints yes write_hbonds yes write_orientations no num_scored_conformers 1 rank_ligands no
Then we run it using dock:
dock6 -i footprint.in -o footprint.out
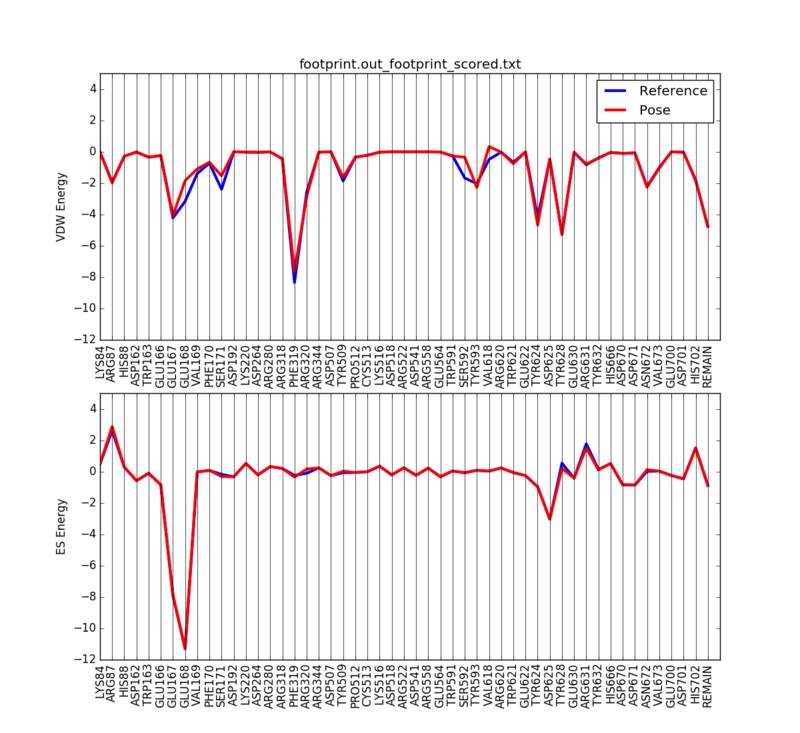
We can then plot the 50 most significant residues using the following python script:
python plot_footprint_single_magnitude.py footprint.out_footprint_scored.txt 50
Docking & Virtual Screening
Now that the ligand and receptor files are set up, we can begin docking the ligand back into the receptor. This makes sure that files are set up correctly, since we should expect the docking results to closely match the crystal structure from the PDB.
Rigid Docking
The first docking method we will be using is rigid docking. With this method, the ligand is kept in a fixed position while the algorithm tries to move/rotate the entire molecule to reduce energy. This is the least computationally intensive method for docking. We can start by creating the input file with the command
vim rigid.in
And using the following parameters.
conformer_search_type rigid use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100 ligand_atom_file 1x70.ligand.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 1x70.ligand.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 2000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes simplex_max_iterations 1000 simplex_tors_premin_iterations 0 simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix rigid.out write_orientations no num_scored_conformers 1 rank_ligands no
Fixed Anchor Docking
In fixed anchor docking, part of the ligand remains fixed, and the rest of it is given some rotational degrees of freedom. This makes it slightly more computationally intensive than rigid docking, but still less intensive than flex docking. So it serves as a middle ground between the two. We can start by creating the input file.
vim fixed.in
Then adding the following parameters.
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 10000 pruning_clustering_cutoff 100 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file ../001.structure/1x70_ligand_wH.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename ../001.structure/1x70_ligand_wH.mol2 use_database_filter no orient_ligand no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1 simplex_trans_step 1 simplex_rot_step 0.1 simplex_tors_step 10 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix 1x70_fad write_orientations no num_scored_conformers 100 write_conformations no cluster_conformations yes cluster_rmsd_threshold 2.0 rank_ligands no
Flexible Docking
In flex docking, the ligand is given full rotational freedom, so the software can put it in whatever conformation it wants. Because of this, flex docking is the most computationally intensive docking method. We can start by creating the input file.
vim flex.in
Then adding the following parameters
conformer_search_type flex write_fragment_libraries no user_specified_anchor no limit_max_anchors no min_anchor_size 5 pruning_use_clustering yes pruning_max_orients 5000 pruning_clustering_cutoff 2500 pruning_conformer_score_cutoff 100.0 pruning_conformer_score_scaling_factor 1.0 use_clash_overlap no write_growth_tree no use_internal_energy yes internal_energy_rep_exp 12 internal_energy_cutoff 100.0 ligand_atom_file 1x70.ligand.min_scored.mol2 limit_max_ligands no skip_molecule no read_mol_solvation no calculate_rmsd yes use_rmsd_reference_mol yes rmsd_reference_filename 1x70.ligand.min_scored.mol2 use_database_filter no orient_ligand yes automated_matching yes receptor_site_file ../002.surface_spheres/selected_spheres.sph max_orientations 1000 critical_points no chemical_matching no use_ligand_spheres no bump_filter no score_molecules yes contact_score_primary no contact_score_secondary no grid_score_primary yes grid_score_secondary no grid_score_rep_rad_scale 1 grid_score_vdw_scale 1 grid_score_es_scale 1 grid_score_grid_prefix ../003.gridbox/grid multigrid_score_secondary no dock3.5_score_secondary no continuous_score_secondary no footprint_similarity_score_secondary no pharmacophore_score_secondary no descriptor_score_secondary no gbsa_zou_score_secondary no gbsa_hawkins_score_secondary no SASA_score_secondary no amber_score_secondary no minimize_ligand yes minimize_anchor yes minimize_flexible_growth yes use_advanced_simplex_parameters no simplex_max_cycles 1 simplex_score_converge 0.1 simplex_cycle_converge 1.0 simplex_trans_step 1.0 simplex_rot_step 0.1 simplex_tors_step 10.0 simplex_anchor_max_iterations 500 simplex_grow_max_iterations 500 simplex_grow_tors_premin_iterations 0 simplex_random_seed 0 simplex_restraint_min no atom_model all vdw_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/vdw_AMBER_parm99.defn flex_defn_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex.defn flex_drive_file /gpfs/projects/AMS536/zzz.programs/dock6.9_release/parameters/flex_drive.tbl ligand_outfile_prefix flex.out write_orientations yes num_scored_conformers 25 write_conformations yes cluster_conformations yes cluster_rmsd_threshold 2 rank_ligands no
Because this method is so intensive, we do not want to run the code on the login node of seawulf. Instead, we should submit a job to run it on one of the compute nodes. To do this, we can create the following slurm script.
vim slurm.sh
Then fill it in with the following commands.
#!/bin/bash #SBATCH --time=3:00:00 #SBATCH --nodes=1 #SBATCH --ntasks=24 #SBATCH --job-name=1x70_flex #SBATCH --output=1x70_flex.txt #SBATCH -p short-24core cd $SLURM_SUBMIT_DIR dock6 -i flex.in -o 1x70_flex.out
It is worth it to note that short-24core processor may not be the best one to use, as other people may be using all of the nodes. You can use the command
sinfo
to see which nodes are currently in use and pick one of the short processors that has some nodes idle. After submitting the slurm job, you can check the status of it using the command
squeue -u netid
If the entry under the st (status) says pd, then that means your code is currently waiting to start. If it says r, then it is currently being run and it will show the elapsed time as well. Once the job has finished, all of the output files should be left in your current directory. You can load the .mol2 file in Chimera to see if it matches your crystal structure.
Virtual Screen
[[File:|thumb|center|800px|image placeholder]]