Cross Docking SB2024 V1 DOCK6.10 A
The purpose of this tutorial is to develop a uniform method to test cross docking across the Rizzo lab with the DOCK software. Note any data in this tutorial is solely for the purpose of example.
Contents
I.Introduction
- Cross docking is a test which is fundamentally similar to pose reproduction. If you are not experienced running pose reproduction yet, begin with:
https://ringo.ams.stonybrook.edu/index.php/Pose_Reproduction_Tutorial
- Cross docking measures pose reproduction accuracy with differing protein conformations/ structures as an additonal variable. It is a more translatable test to "real world" virtual screening, because it tests the ability to identify native poses, even when protein conformation/ sidechain packing is not induced to the particular ligand. When virual screening with a rigid receptor, the particular conformation chosen will not be ideal for all binder chemotypes, but nonetheless it is desirable to predict near native poses.
- The outcomes for cross docking are the same as pose reproduction, although there is a fourth outcome termed "nonviable". This is when a ligand is energetically incompatible in its native pose when complexed in an alternative, rigid, structure of the protein.
II.Necessary files
Scripts to run Cross Docking in batch mode are found at:
https://github.com/rizzolab/Benchmarking_and_Validation
This tutorial rebuilds systems in an aligned frame. Thus standard test set files are not applicable because proteins families will not necessarily be aligned at protein backbone. Instead of directly using a finalized and prepared test set as Pose Reproduction does, this prepares files from initial preparatory files. The list of necessary files are:
${pdb_id}.lig.moe.mol2
${pdb_id}.cof.moe.mol2 (if applicable)
${pdb_id}.rec.foramber.pdb
For further explanation of these 3 files see section "Preliminary File Preparation":
https://ringo.ams.stonybrook.edu/index.php/Test_Set_Tutorial_V1
III.Preparing protein families
Enter CrossDocking Directory:
cd Benchmarking_and_Validation/CrossDocking/
Protein families are a set of structures of the same protein in different conditions. This tutorial will not cover how protein families are determined, although one option is to restrict a family to structures with a single "UniProt ID" and no differing mutations within an active site, and with co-crystal ligands occupying the same active site. Lists of protein families can be found at (Note: see each corresponding paper for how families were determined):
https://ringo.ams.stonybrook.edu/index.php/Rizzo_Lab_Downloads
A list of PDB codes for each protein family needs to be provided:
cd zzz.family_lists
For each protein family, create a file with the name of the protein family, and list all PDB structures for that family:
vi Acetylcholinesterase.txt
The file should like this (Note: The first PDB listed will be the reference which all other proteins are aligned to - a criteria should be chosen for which is first):
1EVE
1GPK
1GPN
...
After creating a file listing PDB codes for each protein family, create a file listing protein family names:
vi zzz.Families.txt
The file should look like this:
Acetylcholinesterase
...
IV.Aligning protein families
The first step is to align protein families using the program UCSF Chimera along the backbone. This is done using the "mmaker" command and aligning all proteins to a single reference. The co-crystal ligand in each structure undergoes the same transformation alignment. (If Chimera is not available as a module this can be done in the gui.)
module load chimera/1.13.1
Certain variables need to be sourced every time a new session is started to run these scripts:
source 000.source.env.sh
Edit slurm header and set path to testset (Same testset for Pose Reproduction which should have zzz.master as a subdirectory):
vi 001.align.submit.sh
sbatch 001.align.submit.sh
The aligned files for ligand (${pdb_id}.lig.moe.mol2) and receptor (${pdb_id}.rec.foramber.pdb) are found in:
cd Alignment/Acetylcholinesterase/mol2/
The visual alignment of each protein family should be checked in Chimera gui:
Statistics on alignment should also be inspected. This can be found in:
Alignment/Acetylcholinesterase/${pdb_id}
In this folder is the alignment data for this particular structure:
vi chimera.out
Below is an example of the output of a structure which has a good alignment to the reference. All pairs were used in the alignment and the RMSD of the 2 structures in the alignment is 0.321 angstrom:
In general a good alignment will include (i) at least 90% of the pairs, with (ii) an RMSD less than 2.0 angstrom for the pairs. Anything outside of this range should be rejected from a protein family.
Below is an example of the output of a structure which has a poor alignment to the reference. Only 20% (108 / 528) of pairs are used in the alignment which produces a low RMSD. The RMSD from all pairs is higher than the 2.0 cutoff. This structure should be removed from the family:
V.System Preparation of Aligned Structures
If this is a new session remember to run:
source 000.source.env.sh
Python2 scripts are required for the next step (Following command should load py/2.7.15 for current seawulf setup):
module load py/
DOCK6 is also used in the next step so load appropriate DOCK6 compilers (Different DOCK compilations have different compilers):
module load intel-stack
Run build scripts:
sbatch 002a.build.submit.sh
While not yet implemented here, bookkeeping scripts and test cases covered in link below would be wise to do. These scripts and test ensure everything ran successfully when converting input files into DOCK6 ready format:
https://ringo.ams.stonybrook.edu/index.php/Test_Set_Tutorial_V1#III.G._Bookkeeping_of_Heavy_Atoms
Now transfer docking files to location where they will be used for docking.
bash 002b.transfer.sh
Any systems which did not successfully create all dock files (if any) will be listed in:
vi Incomplete_Builds.txt
VI.Docking molecules
The next step writes the dock input files for process. As specified in the Pose Reproduction manual, while it is okay to copy the parameters used in this script, a dock input file should be generated interactively so that any potential differences in the DOCK6 version used to create this file (relative to version with tutorial user) will be accounted for, and a version appropriate input file is used. This file would be included in 003a.write.files.sh by using the same variables already included.
bash 003a.write.files.sh
This step writes 2 files:
min.in
flx.in
min.in cartesian minimizes each ligand in each in each receptor starting from its native cognate pose. This tests if a ligand's native pose is energetically compatible with a particular structure. If the RMSD of minimization is greater than 2 angstroms, it is considered a non-viable pairing.
flx.in flexibly docks each ligand into each receptor similar to Pose Reproduction.
To check these files exist:
cd zzz.crossdock/Acetylcholinesterase/1ACJ/1EVE
After docking is complete each directory should have the following files (This is the docking of (ligand 1EVE) into (protein 1ACJ):
cd zzz.crossdock/Acetylcholinesterase/1ACJ/1EVE
1EVE_1ACJ.min_scored.mol2
1EVE_1ACJ.FLX_scored.mol2
Now run the docking. Input slurm header and specify the path to the DOCK6 folder uppermost directory:
vi 003b.run.crossdock.sh
dock_dir="/path/to/dock6"
Again make sure DOCK6 compilers are loaded:
sbatch 003b.run.crossdock.sh
VII.Cross Docking Analysis
Next quantify each outcome into "Success" "Score Fail" "Sample Fail" or "Non viable".
bash 004.results.sh
This creates a file for each system with a numeric code indicating outcome:
zzz.crossdock/Acetylcholinesterase/${pdb_id}.outcomeh.txt
Next reorder each protein family with the ligand having the highest score in all receptors. For each success +3, for each score fail +2, for each sample fail +1. This has the general trend of putting successes towards the top of the list and sample failures towards the bottom.
bash 005.sort.lists.sh
This is written to:
zzz.rearrange_lists/Acetylcholinesterase_Rearrangeh.txt
Do some reformatting of results:
bash 006.rerun.results_h.sh
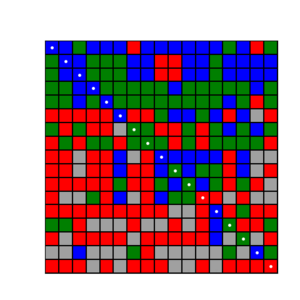
Now make heatmaps:
bash 007.make.heatmaps.sh
The result will be an N x N matrix where N is the size of the crossdocking protein family. The negative diagonal represents cocrystal ligand and protein structures, which we call "cognate pair". Each row is a single ligand docked into every protein structure, and each column is a single protein structure with every ligand docked.
The file is located at:
vi HeatMaps/Acetylcholinesterase.No_PDB.RMSDh.heatmap.png
To list the PDB codes on the axis open the file and uncomment the following lines:
vi zzz.crossdock_scripts/Make_ploth.py
plt.xticks(range(ncols), col_labels, rotation = 90, fontsize = fontsz)
plt.yticks(range(nrows), row_labels, fontsize = fontsz)
-SEE README FILE IN GIT REPO FOR ADDTIONAL DETAILS THAT MAY NOT BE COVERED HERE
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
Tutorial Written By: Christopher Corbo, Rizzo Lab, Stony Brook University (This tutorial was last updated 02/29/2024)
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>