2025 DOCK tutorial 2 with PDBID 1XMU
Contents
Introduction
Structure Preparation
The objectives of this section are:
1. Structural Evaluation
a. Identify any missing loops in the protein structure. b. Assess metal coordination atoms for key interactions with the protein.
2. Protein Receptor Preparation
a. Add hydrogen atoms. b. Assign appropriate charges.
3. Ligand Preparation
a. Add hydrogen atoms. b. Assign appropriate charges.
1. Structural Evaluation
1. Open the downloaded PDB structure in Chimera (In this tutorial, 1XMU). 2. Check for missing loops—these appear as dashed lines in the structure. 3. There are no missing loops in the structure 1XMU, so no further action is needed.
If you find missing loops in your structure, you will need to fix them before proceeding.
Structural Evaluation
1. Open the downloaded PDB structure in Chimera (In this tutorial, 1XMU).
2. Check for missing loops—these appear as dashed lines in the structure.
3. There are no missing loops in the structure 1XMU, so no further action is needed.
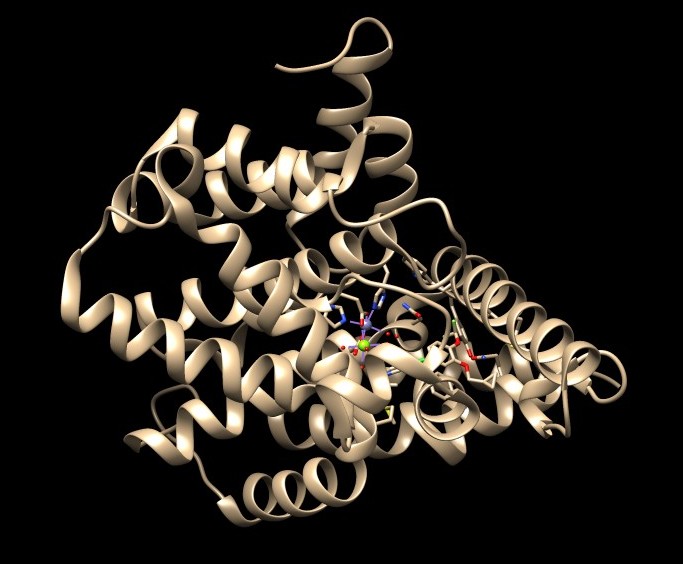 If you find missing loops in your structure, you will need to fix them before proceeding.
If you find missing loops in your structure, you will need to fix them before proceeding.